Units#
The primary data in this dataset is the recorded acrtivity of isolated units. A
number of metrics are used to isolate units through spike sorting, and these
metrics can be used to access how well isolated they are and the quality of
each unit. The units dataframe provides many of these metrics, as well as
parameterization of the waveform for each unit that passed initial QC,
including
firing rate: mean spike rate during the entire session
presence ratio: fraction of session when spikes are present
ISI violations: rate of refractory period violations
Isolation distances: distance to nearest cluster in Mihalanobis space
d’: classification accuracy based on LDA
SNR: signal to noise ratio
Maximum drift: Maximum change in spike depth during recording
Cumulative drift: Cumulative change in spike depth during recording
Accessing the units#
import os
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
import pandas as pd
from allensdk.brain_observatory.ecephys.ecephys_project_cache import EcephysProjectCache
/opt/envs/allensdk/lib/python3.8/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
# Example cache directory path, it determines where downloaded data will be stored
output_dir = '/root/capsule/data/allen-brain-observatory/visual-coding-neuropixels/ecephys-cache/'
manifest_path = os.path.join(output_dir, "manifest.json")
cache = EcephysProjectCache.from_warehouse(manifest=manifest_path)
session_id = 750332458 # An example session id
session = cache.get_session_data(session_id)
1D Waveform features:
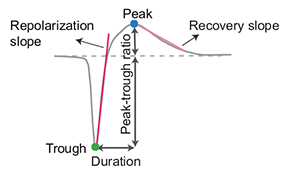
For more information on these:
AllenInstitute/ecephys_spike_sorting AllenInstitute/ecephys_spike_sorting
The units table#
The units table contains important information about each unit that was recorded, including its spike sorting quality metrics, its 3D position in the Allen Common Coordinate Framework, and the structure in which it’s located.
Here is a brief summary of the columns in this table:
column name |
description |
|---|---|
|
the identifier for this unit assigned by the AllenSDK (unique across the entire dataset) |
|
peak-to-trough ratio of the average spike waveform |
|
amplitude (in microvolts) of the average spike waveform |
|
a measure of the approximate fraction of spikes missing from this unit (default threshold = 0.1) |
|
the identifier for this unit assigned by the spike sorting algorithm (unique within each probe) |
|
the integrated distance (in microns) that the unit drifted across the whole session |
|
a measure of how separable this unit’s waveforms are from its neighbors’ |
|
mean spike rate across the whole session |
|
a measure of this unit’s level of contamination (default threshold = 0.5) |
|
a measure of how separable this unit’s waveforms are from its neighbors’ (higher is better) |
|
a measure of how separable this unit’s waveforms are from its neighbors’ (lower is better) |
|
the index of this unit within the probe it was recorded with |
|
the maximum distance (in microns) the unit drifted across the whole session |
|
a measure of this unit’s level of contamination |
|
a measure of the fraction of spike missing from this unit |
|
the identifier for this unit’s peak channel (can be used as an index into the |
|
the fraction of the session over which this unit had spikes detected (default threshold = 0.9) |
|
slope of the waveform between the trough and the peak |
|
slope of the waveform back to 0 after the peak |
|
a measure of this unit’s level of contamination |
|
the ratio of the waveform amplitude relative to the background noise on the peak channel |
|
distance the waveform extends above and below the peak channel |
|
speed of waveform propagation above the peak channel |
|
speed of waveform propagation below the peak channel |
|
time between the waveform peak and trough |
|
filter properties of the probe used to record this unit |
|
local index of this unit’s peak channel |
|
horizontal position of this unit on the probe |
|
identifier of the probe used to record this unit |
|
vertical position of this unit on the probe |
|
CCF region where this unit is located |
|
CCF structure ID where this unit is located |
|
alias for |
|
CCF coordinate along the A/P axis |
|
CCF coordinate along the D/V axis |
|
CCF coordinate along the L/R axis |
|
name of the probe used to record this unit |
|
not used |
|
spike band sampling rate of the probe used to record this unit |
|
LFP band sampling rate of the probe used to record this unit |
|
|
Working with the units table#
Example: Units
Get the units dataframe for this session.
What the the metrics? (i.e. what are the columns for the dataframe?
How many units are there? How many units per structure?
session.units.head()
| waveform_PT_ratio | waveform_amplitude | amplitude_cutoff | cluster_id | cumulative_drift | d_prime | firing_rate | isi_violations | isolation_distance | L_ratio | ... | ecephys_structure_id | ecephys_structure_acronym | anterior_posterior_ccf_coordinate | dorsal_ventral_ccf_coordinate | left_right_ccf_coordinate | probe_description | location | probe_sampling_rate | probe_lfp_sampling_rate | probe_has_lfp_data | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| unit_id | |||||||||||||||||||||
| 951817231 | 0.293351 | 101.641410 | 0.001248 | 8 | 392.48 | 6.461795 | 15.773666 | 0.020093 | 147.423046 | 0.000259 | ... | 8.0 | grey | -1000 | -1000 | -1000 | probeA | See electrode locations | 29999.968724 | 1249.998697 | True |
| 951817222 | 1.427508 | 74.654970 | 0.032535 | 7 | 948.33 | 5.638511 | 6.423025 | 0.007457 | 95.080849 | 0.000727 | ... | 8.0 | grey | -1000 | -1000 | -1000 | probeA | See electrode locations | 29999.968724 | 1249.998697 | True |
| 951817272 | 0.240866 | 182.350545 | 0.000218 | 13 | 578.80 | 4.865528 | 25.891454 | 0.002123 | 121.137882 | 0.017477 | ... | 8.0 | grey | -1000 | -1000 | -1000 | probeA | See electrode locations | 29999.968724 | 1249.998697 | True |
| 951817282 | 0.650177 | 183.182025 | 0.000223 | 14 | 545.47 | 4.402664 | 9.177656 | 0.001370 | 59.655811 | 0.025102 | ... | 8.0 | grey | -1000 | -1000 | -1000 | probeA | See electrode locations | 29999.968724 | 1249.998697 | True |
| 951817316 | 0.387017 | 71.279130 | 0.059431 | 18 | 446.09 | 3.582546 | 10.277127 | 0.050247 | 56.080395 | 0.021113 | ... | 8.0 | grey | -1000 | -1000 | -1000 | probeA | See electrode locations | 29999.968724 | 1249.998697 | True |
5 rows × 40 columns
session.units.columns
Index(['waveform_PT_ratio', 'waveform_amplitude', 'amplitude_cutoff',
'cluster_id', 'cumulative_drift', 'd_prime', 'firing_rate',
'isi_violations', 'isolation_distance', 'L_ratio', 'local_index',
'max_drift', 'nn_hit_rate', 'nn_miss_rate', 'peak_channel_id',
'presence_ratio', 'waveform_recovery_slope',
'waveform_repolarization_slope', 'silhouette_score', 'snr',
'waveform_spread', 'waveform_velocity_above', 'waveform_velocity_below',
'waveform_duration', 'filtering', 'probe_channel_number',
'probe_horizontal_position', 'probe_id', 'probe_vertical_position',
'structure_acronym', 'ecephys_structure_id',
'ecephys_structure_acronym', 'anterior_posterior_ccf_coordinate',
'dorsal_ventral_ccf_coordinate', 'left_right_ccf_coordinate',
'probe_description', 'location', 'probe_sampling_rate',
'probe_lfp_sampling_rate', 'probe_has_lfp_data'],
dtype='object')
How many units are in this session?
session.units.shape[0]
902
Which areas (structures) are they from?
print(session.units.ecephys_structure_acronym.unique())
['grey' 'VISam' 'VISpm' 'VISp' 'IntG' 'IGL' 'LGd' 'CA3' 'DG' 'CA1' 'VISl'
'VISal' 'VISrl']
How many units per area are there?
session.units.ecephys_structure_acronym.value_counts()
grey 558
VISal 71
VISp 63
VISam 60
VISrl 44
VISl 38
VISpm 19
CA1 16
CA3 15
DG 7
IGL 5
LGd 4
IntG 2
Name: ecephys_structure_acronym, dtype: int64
Example: Select ‘good’ units
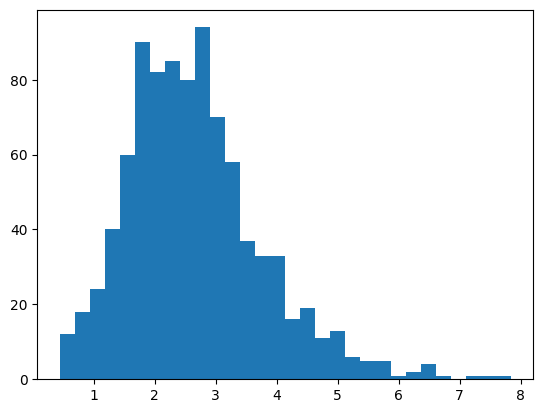
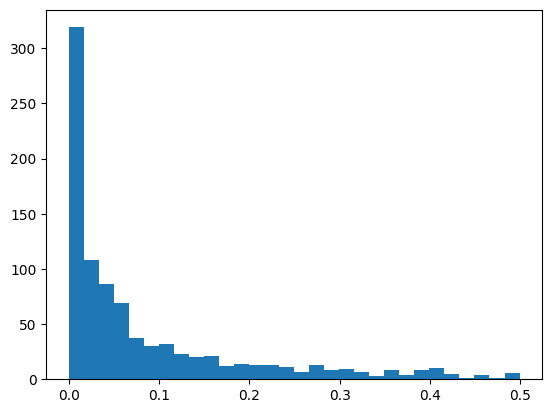
A default is to include units that have a SNR greater than 1 and ISI violations less than 0.5 Plot a histogram of the values for each of these metrics? How many units meet these criteria? How many per structure?
plot a histogram for SNR
plt.hist(session.units.snr, bins=30);
plot a histogram for ISI violations
plt.hist(session.units.isi_violations, bins=30);
good_units = session.units[(session.units.snr>1)&(session.units.isi_violations<0.5)]
len(good_units)
868
good_units.ecephys_structure_acronym.value_counts()
grey 548
VISal 64
VISp 62
VISam 60
VISrl 37
VISl 34
VISpm 18
CA3 15
CA1 15
DG 7
IGL 4
LGd 3
IntG 1
Name: ecephys_structure_acronym, dtype: int64
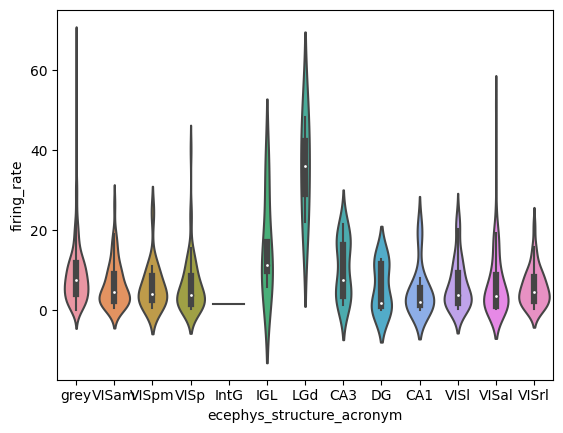
Example: Compare the firing rate of good units in different structures
Make a violinplot of the overall firing rates of units across structures.
import seaborn as sns
sns.violinplot(y='firing_rate', x='ecephys_structure_acronym',data=good_units)
<AxesSubplot:xlabel='ecephys_structure_acronym', ylabel='firing_rate'>
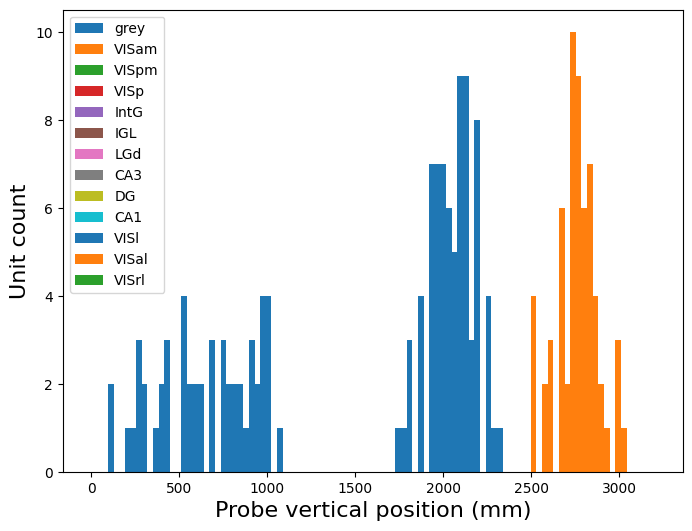
Example: Plot the location of the units on the probe
Color each structure a different color. What do you learn about the vertical position values?
plt.figure(figsize=(8,6))
# restrict to one probe
probe_id = good_units.probe_id.values[0]
probe_units = good_units[good_units.probe_id==probe_id]
for structure in good_units.ecephys_structure_acronym.unique():
plt.hist(
probe_units[probe_units.ecephys_structure_acronym==structure].probe_vertical_position.values,
bins=100, range=(0,3200), label=structure
)
plt.legend()
plt.xlabel('Probe vertical position (mm)', fontsize=16)
plt.ylabel('Unit count', fontsize=16)
plt.show()
Spike Times#
The primary data in this dataset is the recorded acrtivity of isolated units.
The spike times is a dictionary of spike times for each units in the session.
Example: Spike Times
Next let’s find the spike_times for these units.
spike_times = session.spike_times
What type of object is this?
type(spike_times)
dict
How many items does it include?
len(spike_times)
902
len(session.units)
902
What are the keys for this object?
list(spike_times.keys())[:5]
[951817566, 951817557, 951818568, 951818561, 951818553]
These keys are unit ids. Use the unit_id for the first unit to get the spike times for that unit. How many spikes does it have in the entire session?
spike_times[session.units.index[0]]
array([3.79596714e+00, 3.81646716e+00, 3.84250052e+00, ...,
9.75020103e+03, 9.75023709e+03, 9.75027469e+03])
print(len(spike_times[session.units.index[0]]))
153738
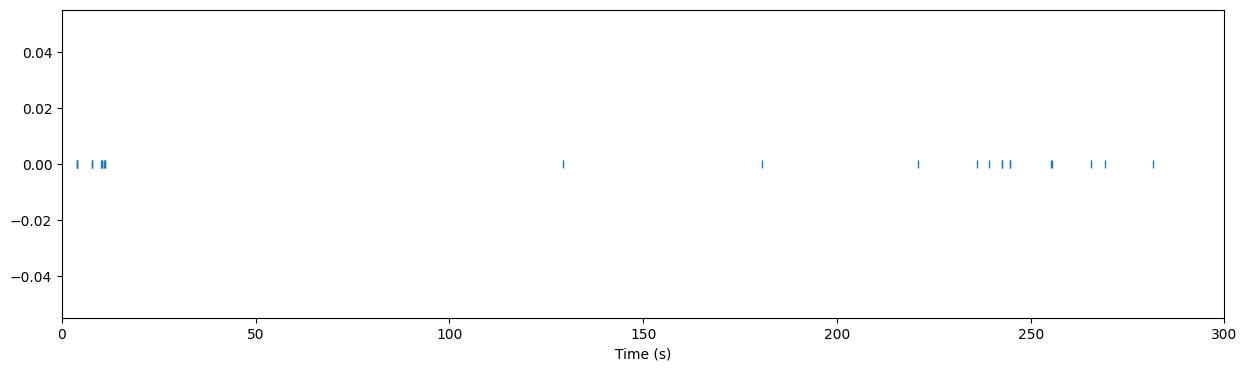
Example: Get the spike times for the units in V1
Use the units dataframe to identify units in ‘VISp’ and use the spike_times to get their spikes. Start just getting the spike times for the first unit identified this way. Plot a raster plot of the spikes during the first 5 minutes (300 seconds) of the experiment.
session.units[session.units.ecephys_structure_acronym=='VISp'].head()
| waveform_PT_ratio | waveform_amplitude | amplitude_cutoff | cluster_id | cumulative_drift | d_prime | firing_rate | isi_violations | isolation_distance | L_ratio | ... | ecephys_structure_id | ecephys_structure_acronym | anterior_posterior_ccf_coordinate | dorsal_ventral_ccf_coordinate | left_right_ccf_coordinate | probe_description | location | probe_sampling_rate | probe_lfp_sampling_rate | probe_has_lfp_data | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| unit_id | |||||||||||||||||||||
| 951814973 | 0.599534 | 26.217945 | 0.014322 | 361 | 345.26 | 2.927932 | 1.503416 | 0.410801 | 61.132264 | 0.001457 | ... | 385.0 | VISp | -1000 | -1000 | -1000 | probeC | See electrode locations | 29999.996461 | 1249.999853 | True |
| 951814989 | 0.366990 | 43.292340 | 0.026720 | 363 | 386.49 | 5.082127 | 1.885503 | 0.000000 | 89.588334 | 0.001691 | ... | 385.0 | VISp | -1000 | -1000 | -1000 | probeC | See electrode locations | 29999.996461 | 1249.999853 | True |
| 951816812 | 0.394103 | 103.924470 | 0.002891 | 561 | 146.34 | 4.708563 | 2.415644 | 0.046132 | 80.151626 | 0.000185 | ... | 385.0 | VISp | -1000 | -1000 | -1000 | probeC | See electrode locations | 29999.996461 | 1249.999853 | True |
| 951815078 | 0.329370 | 111.679035 | 0.051784 | 373 | 165.93 | 3.799649 | 4.062188 | 0.100211 | 55.356604 | 0.010204 | ... | 385.0 | VISp | -1000 | -1000 | -1000 | probeC | See electrode locations | 29999.996461 | 1249.999853 | True |
| 951815150 | 0.526429 | 232.830975 | 0.001259 | 382 | 302.18 | 6.045597 | 0.516495 | 0.000000 | 80.501412 | 0.000006 | ... | 385.0 | VISp | -1000 | -1000 | -1000 | probeC | See electrode locations | 29999.996461 | 1249.999853 | True |
5 rows × 40 columns
unit_id = session.units[session.units.ecephys_structure_acronym=='VISp'].index[12]
spikes = spike_times[unit_id]
plt.figure(figsize=(15,4))
plt.plot(spikes, np.repeat(0,len(spikes)), '|')
plt.xlim(0,300)
plt.xlabel("Time (s)")
Text(0.5, 0, 'Time (s)')
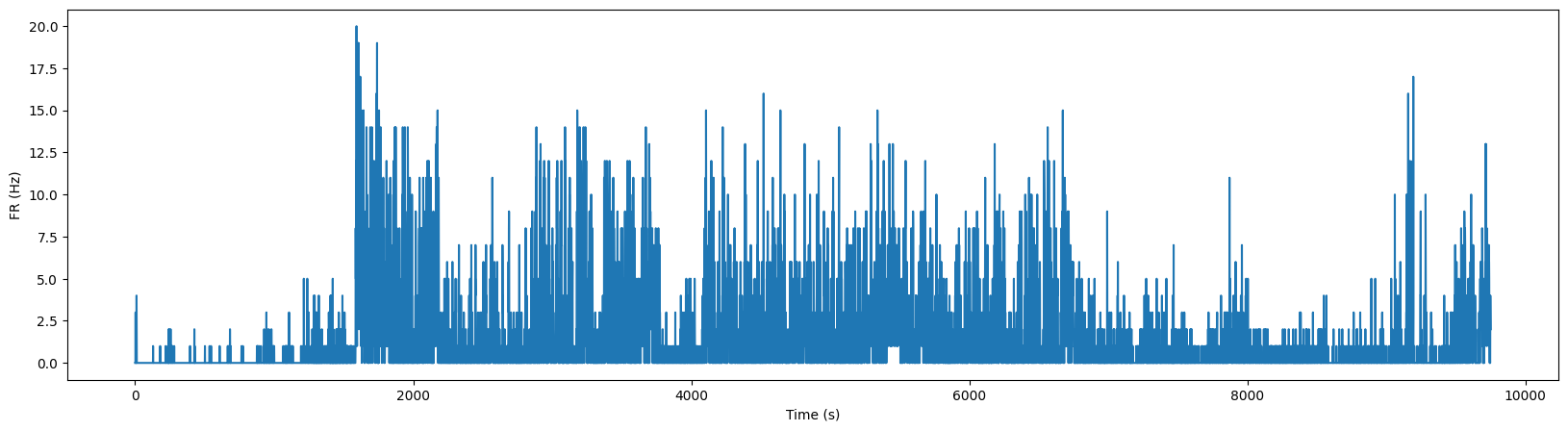
Example: Plot the firing rate for this units across the entire session
A raster plot won’t work for visualizing the activity across the entire session as there are too many spikes! Instead, bin the activity in 1 second bins.
numbins = int(np.ceil(spikes.max()))
binned_spikes = np.empty((numbins))
for i in range(numbins):
binned_spikes[i] = len(spikes[(spikes>i)&(spikes<i+1)])
plt.figure(figsize=(20,5))
plt.plot(binned_spikes)
plt.xlabel("Time (s)")
plt.ylabel("FR (Hz)")
Text(0, 0.5, 'FR (Hz)')
Example: Plot firing rates for units in V1
Now let’s do this for up to 50 units in V1. Make an array of the binned activity of all units in V1 called ‘v1_binned’. We’ll use this again later.
v1_units = session.units[session.units.ecephys_structure_acronym=='VISp']
numunits = len(v1_units)
if numunits>50:
numunits=50
v1_binned = np.empty((numunits, numbins))
for i in range(numunits):
unit_id = v1_units.index[i]
spikes = spike_times[unit_id]
for j in range(numbins):
v1_binned[i,j] = len(spikes[(spikes>j)&(spikes<j+1)])
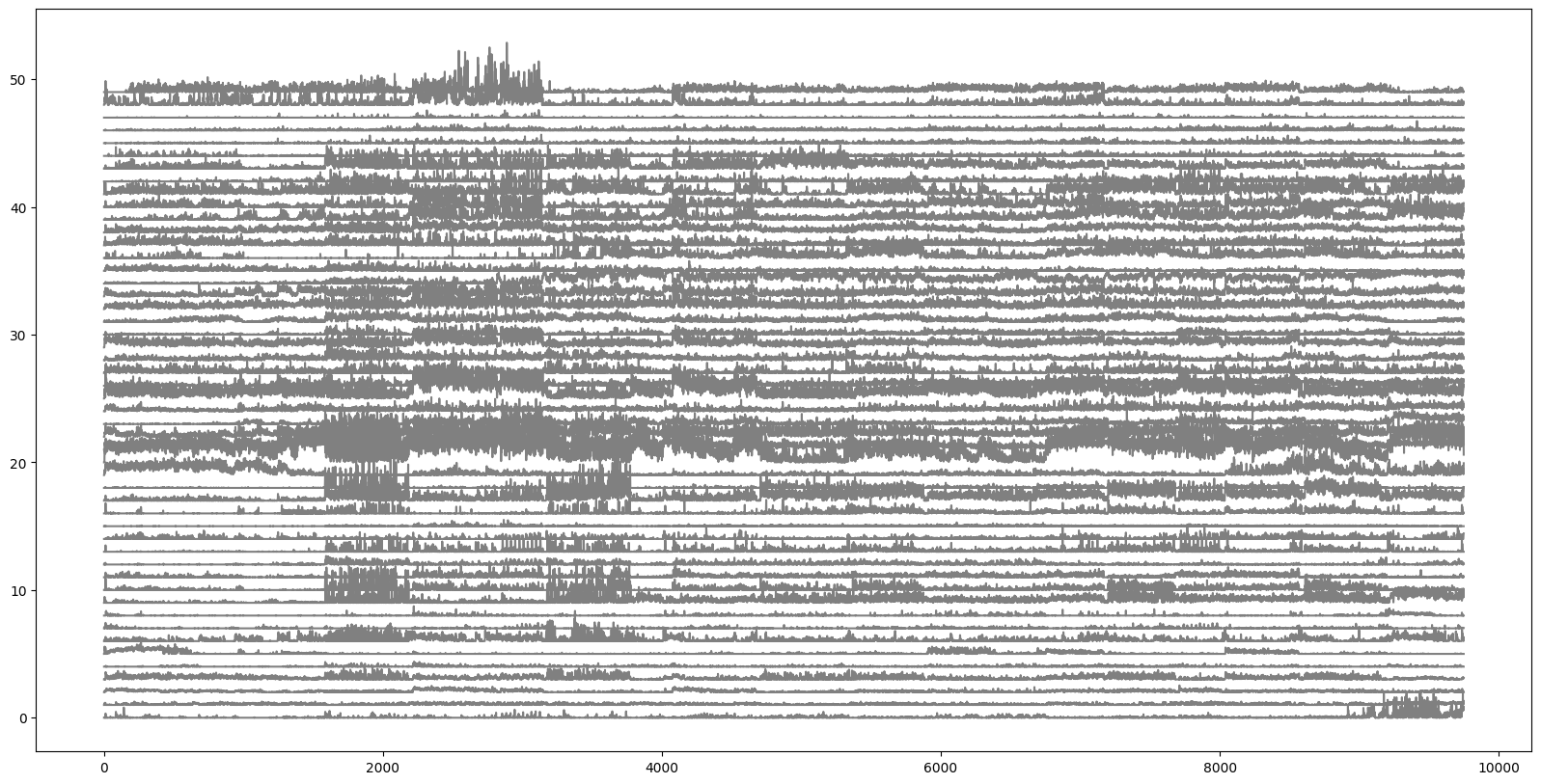
Plot the activity of all the units, one above the other
plt.figure(figsize=(20,10))
for i in range(numunits):
plt.plot(i+(v1_binned[i,:]/30.), color='gray')
Unit waveforms#
For each unit, the average action potential waveform has been recorded from each
channel of the probe. This is contained in the mean_waveforms object. This is
the characteristic pattern that distinguishes each unit in spike sorting, and it
can also help inform us regarding differences between cell types.
We will use this in conjuction with the channel_structure_intervals function
which tells us where each channel is located in the brain. This will let us get
a feel for the spatial extent of the extracellular action potential waveforms in
relation to specific structures.
Example: Unit waveforms
Get the waveform for one unit.
waveforms = session.mean_waveforms
What type of object is this?
type(waveforms)
dict
What are the keys?
list(waveforms.keys())[:5]
[951817566, 951817557, 951818568, 951818561, 951818553]
Get the waveform for one unit
unit = session.units.index.values[400]
wf = session.mean_waveforms[unit]
What type of object is this? What is its shape?
type(wf)
xarray.core.dataarray.DataArray
wf.coords
Coordinates:
* channel_id (channel_id) int64 850176026 850176028 ... 850176790 850176792
* time (time) float64 0.0 3.333e-05 6.667e-05 ... 0.002667 0.0027
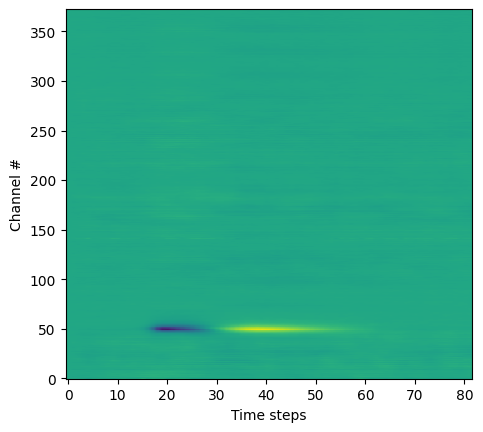
wf.shape
(373, 82)
plt.imshow(wf, aspect=0.2, origin='lower')
plt.xlabel('Time steps')
plt.ylabel('Channel #')
Text(0, 0.5, 'Channel #')
Example: Unit waveforms
Use the channel_structure_intervals to get information about where each
channel is located.
We need to pass this function a list of channel ids, and it will identify channels that mark boundaries between identified brain regions.
We can use this information to add some context to our visualization.
# pass in the list of channels from the waveforms data
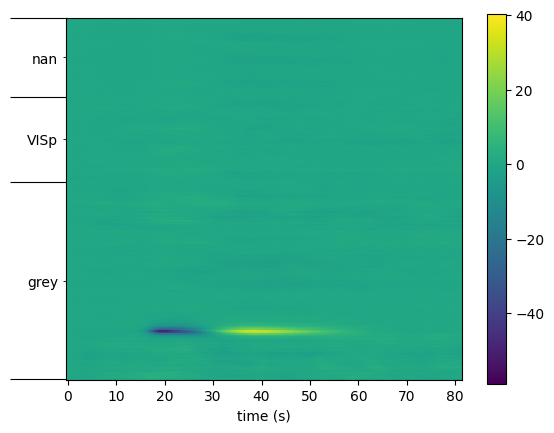
ecephys_structure_acronyms, intervals = session.channel_structure_intervals(wf.channel_id.values)
print(ecephys_structure_acronyms)
print(intervals)
['grey' 'VISp' nan]
[ 0 204 292 373]
Place tick marks at the interval boundaries, and labels at the interval midpoints.
fig, ax = plt.subplots()
plt.imshow(wf, aspect=0.2, origin='lower')
plt.colorbar(ax=ax)
ax.set_xlabel("time (s)")
ax.set_yticks(intervals)
# construct a list of midpoints by averaging adjacent endpoints
interval_midpoints = [ (aa + bb) / 2 for aa, bb in zip(intervals[:-1], intervals[1:])]
ax.set_yticks(interval_midpoints, minor=True)
ax.set_yticklabels(ecephys_structure_acronyms, minor=True)
plt.tick_params("y", which="major", labelleft=False, length=40)
plt.show()
Let’s see if this matches the structure information saved in the units table:
session.units.loc[unit, "ecephys_structure_acronym"]
'grey'
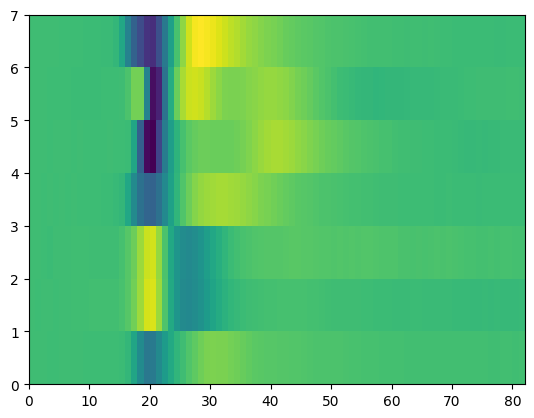
Example: Plot the mean waveform for the peak channel for each unit in the dentate gyrus (DG)
Start by plotting the mean waveform for the peak channel for the unit we just looked at. Then do this for all the units in DG, making a heatmap of these waveforms
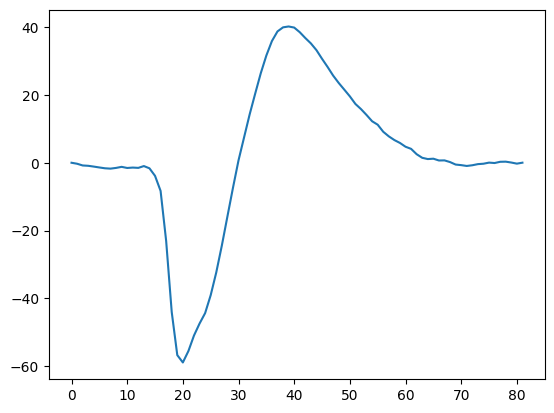
Find the peak channel for this unit, and plot the mean waveform for just that channel
channel_id = session.units.loc[unit, 'peak_channel_id']
print(channel_id)
850176126
plt.plot(wf.loc[{"channel_id": channel_id}])
[<matplotlib.lines.Line2D at 0x7fc902f50f40>]
fig, ax = plt.subplots()
th_unit_ids = good_units[good_units.ecephys_structure_acronym=="DG"].index.values
peak_waveforms = []
for unit_id in th_unit_ids:
peak_ch = good_units.loc[unit_id, "peak_channel_id"]
unit_mean_waveforms = session.mean_waveforms[unit_id]
peak_waveforms.append(unit_mean_waveforms.loc[{"channel_id": peak_ch}])
time_domain = unit_mean_waveforms["time"]
peak_waveforms = np.array(peak_waveforms)
plt.pcolormesh(peak_waveforms)
<matplotlib.collections.QuadMesh at 0x7fc9056a42e0>