Transgenic tools#
There are a large number of transgenic tools available that enable scientists to visualize, record, and manipulate cells in the mouse. We will briefly describe the general transgenic approaches that we use here, but this is not an exhaustive explanation of this full space.
Cre/lox recombination approaches#
The Cre/lox recombination system is a conditional approach that allows genes to be removed or expressed within specific tissues or cell types. Cre is a site-specific recombinase that drives the recombination of DNA specifically at lox-P sites. Engineering loxP on either side of a gene of interest in a cell that expresses Cre will result in that gene being removed from that cell. This technique can be used to induce the expression of a gene of interest by having a loxP-STOP-loxP sequence in front of the gene of interest. Without Cre, this gene will not be expressed due to the STOP sequence. But when Cre is present, the STOP sequence is removed, and the gene will be expressed. This is how we have used Cre lines in these experiments to drive the expression of chosen reporters.
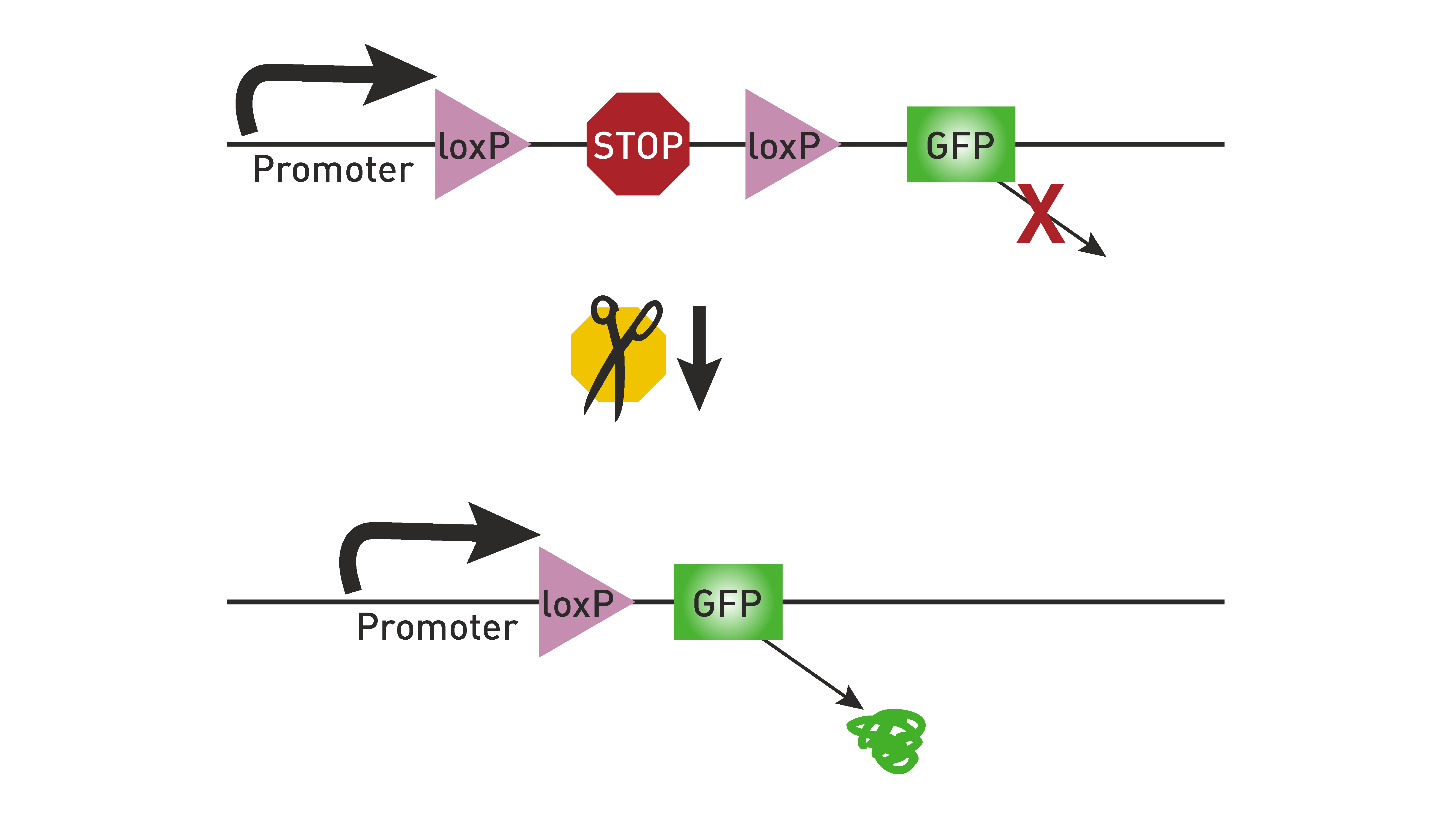
Fig. 2 Excising a STOP sequence with Cre induces expression of the gene of interest. Image taken from The Jackson Laboratory. :::#
Genes of interest can be inserted into the mouse with the use of viruses, or the creation of transgenic mouse lines that have the gene inserted into their genome. A common technique for using transgenic mice is to create separate driver lines and reporter lines, then crossing them together in order to get the desired gene into the desired brain region/cell type/etc. This is much more efficient than making making a separate line for every possible gene to be directly expressed in every possible cell type: driver and reporter lines allow us to “mix and match” and create the exact combination we need for a given experiment.

Fig. 3 Crossing a driver line with a reporter line results in offspring expressing the reporter in a subset of cells defined by the driver line. Image taken from The Jackson Laboratory.#
Driver lines used here#
A driver line is a transgenic mouse line in which mice have had the gene for Cre or another recombinase (such as tTA) inserted into their genome, commonly under the control of a specific promoter. Promoters are often expressed pretty broadly across the brain, but can also be extremely specific. Here are the driver lines and their expression patterns for lines used in our data sets. Note that in the absence of a reporter, Cre on its own does not greatly affect the mouse’s physiology.
Name |
Description |
MGI ID |
Datasets |
|---|---|---|---|
Adora2a-Cre |
In striatum, drives expression in medium spiny neurons (MSNs) expressing the D2 dopamine receptor. These are MSNs part of the indirect pathway. |
MGI:3852493 |
Cell Type Lookup Table |
Camk2a-tTA |
A broadly expressed promoter that uses the tetracycline-controlled transactivator protein (tTA) to drive the expression of reporters under the TRE or tetO element. In many of our applications, we combine both Cre and tTA to drive the expression of our reporters. |
BCI, Visual Coding 2P, Visual Behavior 2P, V1 Deep Dive |
|
Cart-IRES2-Cre |
Targets Cocaine- and amphetamine-regulated transcript gene expressing neurons. This gene encodes a preproprotein that is proteolytically processed to generate multiple biologically active peptides. These peptides play a role in appetite, energy balance, maintenance of body weight, reward and addiction, and the stress response. This line was chosen to label neurons in the medial habenula and the paraventricular nucleus of the thalamus. |
MGI:5704439 |
Single Cell Morphology |
Chat-IRES-Cre-neo |
Drives expression in cholinergic neurons. |
||
Cux2-CreERT2 |
In cortex, drives expression in excitatory neurons in layer 2/3 and 4. |
MGI:5014172 |
Visual Coding 2P |
Dbh-Cre-KI |
Labels a small population of ~1200 neurons in the Pons (per hemisphere) that release norepinephrine to targets brainwide (except striatum). |
MGI:6305870 |
Single Cell Morphology |
Drd1a-Cre |
In striatum, drives expression in medium spiny neurons (MSNs) expressing the D1 dopamine receptor. These are MSNs part of the direct pathway. |
Cell Type Lookup Table |
|
Emx1-IRES-Cre |
In cortex, a pan-excitatory driver - drives expression in excitatory neurons across all layers. Emx1-IRES-Cre;Camk2a-tTA;Ai93 and Emx1-IRES-Cre;Camk2a-tTA;Ai94 mice were found to exhibit inter-ictal events suggesting that the dense expression of GCaMP6 throughout development could be disrupting normal physiological activity. [Steinmetz et al., 2017] |
MGI:2684610 |
Visual Coding 2P |
Fezf2-CreER |
In cortex, drives expression in corticofugal excitatory neurons in layer 5. |
MGI:6720836 |
Visual Coding 2P |
Gal-Cre_KI187 |
Targets neurons that express the neuropeptide Galanin, chosen to label neurons in midline and interlaminar nuclei in the thalamus. Also targets cells in the hypothalamus, pons and medulla. |
MGI:4367008 |
Single Cell Morphology |
Nr5a1-Cre |
In cortex, drives expression in excitatory neurons in layer 6. |
Visual Coding 2P |
|
Ntsr1-Cre_GN220 |
In cortex, drives expression in a sub-population of excitatory neurons in layer 4. |
MGI:3836636 |
Visual Coding 2P |
Pvalb-IRES-Cre |
Drives expression in Parvalbumin inhibitory interneurons. |
MGI:3590684 |
Visual Coding 2P, Visual Coding Neuropixels, Visual Behavior Neuropixels |
Rbp4-Cre_KL100 |
In cortex, drives expression in excitatory neurons in layer 5. |
MGI:4367067 |
Visual Coding 2P |
Rorb-IRES2-Cre |
In cortex, drives expression in a sub-population of excitatory neurons in layer 4. |
MGI:5507855 |
Visual Coding 2P |
Scnn1a-Tg3-Cre |
In cortex, drives expression in a sub-population of excitatory neurons in layer 4. Only found in primary sensory areas (e.g. VISp). |
MGI:3850187 |
Visual Coding 2P |
Sim1-Cre-KJ18 |
In cortex, drives expression in cortico-subcortical projecting excitatory neurons in layer 5. |
NP Ultra & Psychedelics |
|
Slc17a6-IRES-Cre |
Labels excitatory neurons with broad expression across most thalamic nuclei, hypothalamus, midbrain, and brainstem, but with weak expression in cortex. |
MGI:6758055 |
BCI |
Slc17a7-IRES2-Cre |
In cortex, a pan-excitatory driver - drives expression in excitatory neurons across all layers. Imaged here in layer 2/3, 4, and 5. |
MGI:5507862 |
Visual Coding 2P, Visual Behavior 2P, V1 Deep Dive |
Sst-IRES-Cre |
Drives expression in Somatostatin inhibitory interneurons. |
Visual Coding 2P, Visual Behavior 2P, Visual Coding Neuropixels, Visual Behavior Neuropixels |
|
Tlx3-Cre_PL56 |
In cortex, drives expression in cortico-cortical projecting excitatory neurons in layer 5. |
MGI:5311700 |
Visual Coding 2P, NP Ultra and Psychedelics |
Vip-IRES-Cre |
Drives expression in Vasoactive Intestinal Peptide inhibitory interneurons. |
Visual Coding 2P, Visual Behavior 2P, Visual Coding Neuropixels, Visual Behavior Neuropixels |
Reporter lines used here#
A reporter line is a transgenic mouse line in which mice have had the gene for a reporter inserted into their genome, with a lox-STOP-lox preceding it so that expression does not occur in the absence of Cre. Common reporters are fluorescent proteins (like GFP), opsins (like ChR2), or calcium indicators (like GCaMP). Here are the reporter lines used in our datasets:
Shorthand |
Name |
Description |
MGI ID |
Datasets |
|---|---|---|---|---|
Ai93 |
TITL-GCaMP6f-D |
Cre/Tet dependent fluorescent GCaMP6f indicator expressing GCaMP6 fast. This is often used alongside Camk2a-tTA to enhance the expression in excitatory neurons. |
MGI:5558086 |
Visual Coding 2P, Visual Behavior 2P |
Ai94 |
TITL-GCaMP6s;Rosa26-ZtTA |
Cre/Tet dependent fluorescent GCaMP6s indicator expressing GCaMP6 slow. This is often used alongside Camk2a-tTA to enhance the expression in excitatory neurons. |
MGI:5607576 |
Visual Coding 2P, V1 Deep Dive |
Ai148 |
TIT2L-GC6f-ICL-tTA2_D |
Cre/Tet dependent fluorescent GCaMP6f indicator expressing GCaMP6 fast. This is a second generation reporter that uses the TIGRE2 construct that contains more tTA to drive higher expression. |
MGI:5904003 |
Visual Coding 2P, Visual Behavior 2P |
Ai162 |
TIT2L-GC6s-ICL-tTA2_D |
Cre/Tet dependent fluorescent GCaMP6s indicator expressing GCaMP6 slow. This is a second generation reporter that uses the TIGRE2 construct that contains more tTA to drives higher expression. |
MGI:6151062 |
Visual Coding 2P |
Ai166 |
TIT2L-MORF-ICL-tTA2 |
A Cre dependent reporter that drives sparse labeling with GFP. The MORF introduces a stochastic translational switch, only labeling 1-5% of Cre+ neurons. |
MGI:6441963 |
Single Cell Morphology |
Ai32 |
Rosa-CAG-LSL-ChR2(H134R)-EYFP-WPRE |
Cre dependent expression of channelrhodopsin-2 (with a gain of function H134R substitution) fused to enhanced yellow fluorescent protein (EYFP) for visualization. Cells expressing ChR2(H134R) are rapidly depolarized by illumination with blue light (450-490 nm). |
MGI:5013789 |
Visual Coding Neuropixels, Visual Behavior Neuropixels, NP Ultra & Psychedelics |
TetO-GCaMP6s |
TetO-GCaMP6s |
MGI:5553332 |
V1 Deep Dive |
|
Ai167 |
TIT2L-ChrimsonR-tdTomato-ICL-tTA2-WPRE |
Cre dependent expression of ChrimsonR fused to enhanced red fluorescent protein (tdTomato) for visualization. Cells expressing ChrimsonR are rapidly depolarized by illumination with red light (620-750 nm). |
MGI:7712128 |
NP Ultra & Psychedelics |
Ai230 |
TIT2L-XCaMPG-WPRE-ICL-ChRmine-oScarlet-IRES2-tTA2-WPRE |
Cre dependent expression of ChRmine fused to red fluorescent protein (oScarlet) with the capacity for dual ChRmine-oScarlet/XCaMP-G reporting in the presence of Cre and tTA |
MGI:7482009 |
BCI |
Viruses used here#
Another technique for delivering transgenes to a mouse is to use viruses, typically adeno-associated viruses (AAVs). The gene of interest is inserted into the AAV genome. When a mouse is injected with the virus, the inserted gene is delivered to infected cells. Viruses are commonly used together with driver or reporter lines, though the recently developed enhancer viruses allow delivery of transgenes to specific cell populations even in wild-type mice. Here are the viruses used in our data sets:
Traditional AAVs#
These AAVs can be paired with driver or reporter lines to drive expression of the gene of interest. In our specific data sets, we injected viruses containing floxed (flanked by loxp) genes into mouse driver lines, ensuring that expression only takes place if the gene infects a cell that is already expressing Cre.
Name |
Description |
Datasets |
|---|---|---|
AAV2-Syn-Flex-ChrimsonR-tdTomato |
Cre dependent expression of ChrimsonR, fused with tdTomato for visualization. Cells expressing ChrimsonR are rapidly depolarized by illumination with red light (590 nm peak). |
Cell Type Lookup Table |
pAAV-Ef1a-DIO-ChRmine-mScarlet-WPRE |
Cre dependent expression of ChRmine, fused with mScarlet for visualization. Cells expressing ChRmine are depolarized by illumination with yellow light (520 nm peak). |
Cell Type Lookup Table |
AAV5-hSyn-DIO-somBiPOLES-mCerulean |
Cre dependent expression of BiPOLES, fused with mCerulean for visualization. BiPOLES is a fusion of ChrminsonR and GtACR, meaning cells expressing it are depolarized by red light and hyperpolarized by blue light. |
Cell Type Lookup Table |
AAV-PHP-eB_Syn-Flex-2xTRE-tTA |
Cre dependent tTA promoter. Often this is used to regulate the gain of expression of Cre/Tet dependent reporters - e.g. to get sparse but strong labeling. |
Single Cell Morphology |
AAV-PHP-eB-7xTRE-3x-GFP |
A Cre/Tet dependent reporter that expresses GFP. |
Single Cell Morphology |
AAV-PHP-eB-7xTRE-TdTomato |
A Cre/Tet dependent reporter that expresses TdTomato, a red fluorescent protein. |
Single Cell Morphology |
pAAV-hSyn1-RiboL1-GCaMP8s-WPRE |
A reporter that drives expression of GCaMP8s calcium indicator in soma |
BCI |
pAAV-CaMK11a-ChRmine-oScarlet-Kv2.1-WPRE |
CaMK11a driven expression of channelrhodopsin ChRmine fused with oScarlet for visualization |
BCI |
Enhancer AAVs#
These AAVs deliver genes together with an enhancer sequence. These enhancers have been selected based on their abundance in specific cell types: as such, the gene will only expressed if the virus infects a cell that makes use of this specific enhancer. This allows us to deliver genes directly to a specific cell type without needing to use a driver line.
name |
description |
datasets |
|---|---|---|
rAAV-3xcore2_eHGT_779m-minBG-CoChR-EGFP-WPRE3-BGHpA |
D1 enhancer - CoChR. Drives expression of CoChR in direct pathway MSNs, fused with GFP for visualization. |
Cell Type Lookup Table |
rAAV-3xcore2_eHGT_445h-minBG-CoChR-EGFP-WPRE3-BGHpA |
D2 enhancer - CoChR. Drives expression of CoChR in indirect pathway MSNs, fused with GFP for visualization. |
Cell Type Lookup Table |
rAAV-3xcore2_eHGT_452h-minBG-CoChR-EGFP-WPRE3-BGHpA |
D2 enhancer - CoChR |
Drives expression of |
CoChR in indirect pathway MSNs, fused with GFP for visualization. |
Cell Type Lookup Table |
Do transgenic tools impact physiology or behavior?#
Using transgenic tools to record or manipulate activity rests on an assumption that the expression of these tools does not greatly impact the underlying physiology or behavior that is being studied. Is this a fair assumption? Largely yes, but not 100%.
The exogenous expression of proteins have been shown to have neurotoxic effects. One notable example is the occurrence of aberrant electrical events in (primarily) Emx1-IRES-Cre;Camk2a-tTA;Ai93 mice that resemble interictal events. These are large, concerted electrical events that spread across most of cortex. As putative interictal events, these might be associated with epileptiform seizures, though they themselves are not seizures. Indeed, no other aberrant behavior or activity was reported in these mice. And while the mechanism of why these mice exhibit these events is unclear, it is likely related to very broad exogenous expression of tTA and GCaMP6f starting very early in development. [Steinmetz et al., 2017]
We further examined whether transgenic expression of GCaMP6 effect physiology this was by performing Neuropixels recordings from mice expressing GCaMP6f under the control of several of the driver lines used in the Visual Coding 2P dataset. The question here is whether the exogenous expression of these proteins (Cre, tTA, and GCaMP6f) in particular neurons would alter the underlying physiology of the population. We compared recordings from Vip-IRES-Cre;Ai148, Sst-IRES-Cre;Ai148, Cux2-CreERT2;Camk2a-tTA;Ai93, and Slc17a7-IRES2-Cre;Camk2a-tTA;Ai93 with those from wild-type mice. Comparing unit yield, firing rate, burst fractions, responsiveness, and lifetime sparseness, we did not find significant differences between these populations. [Siegle et al., 2021]
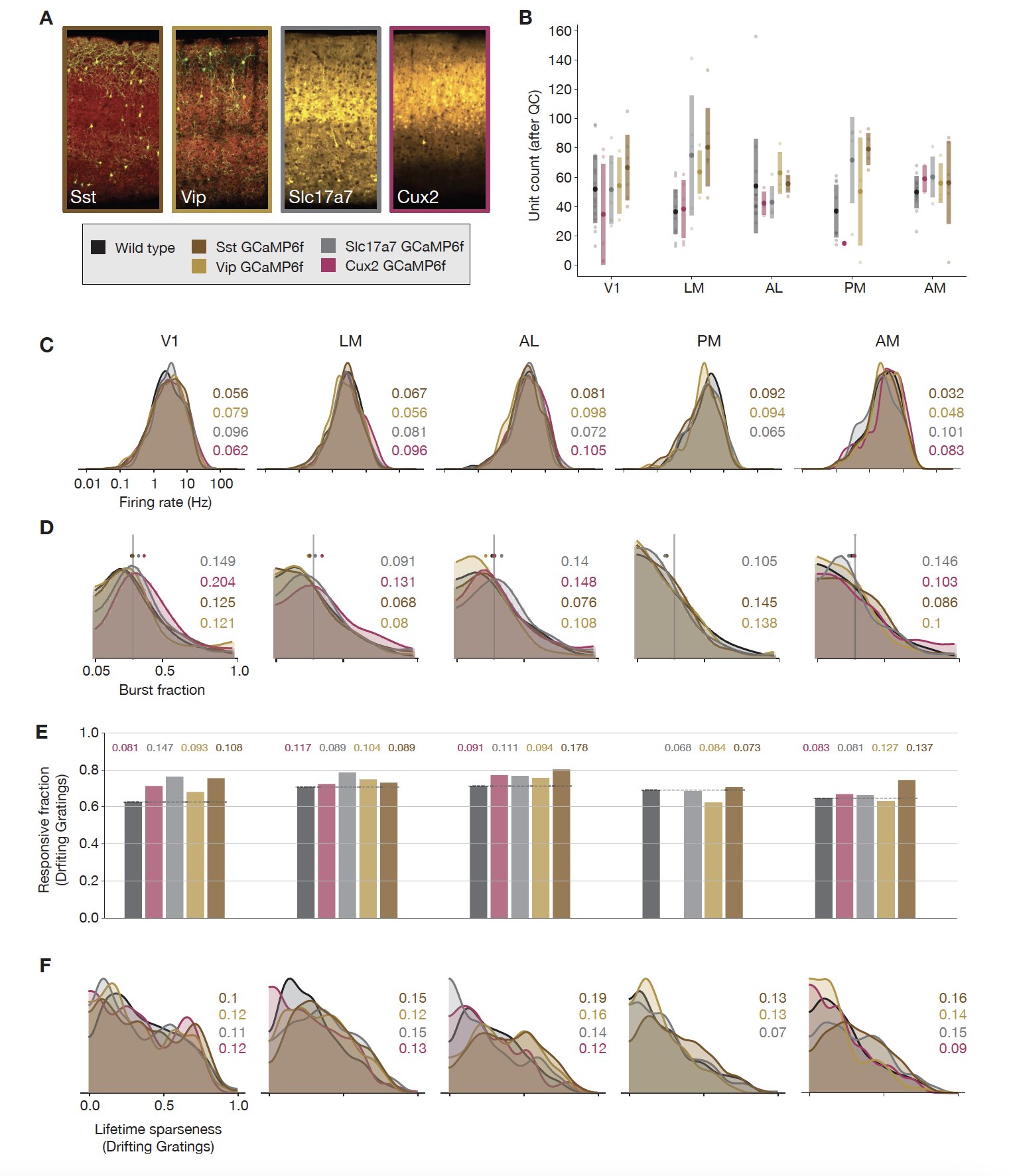
Fig. 4 Comparing responses across GCaMP-expressing mouse lines. (A) GCaMP expression patterns for the four lines used for ephys experiments. (B) Unit yield (following QC filtering) for five areas and five genotypes. Error bars represent standard deviation across experiments; each dot represents a data point from one experiment. (C) Distribution of firing rates for neurons from each mouse line, aggregated across experiments. (D) Distribution of burst fraction (fraction of all spikes that participate in bursts) for neurons from each mouse line, aggregated across experiments. Dots represent the median of each distribution, shown in relation to a reference value of 0.3. (E) Fraction of neurons deemed responsive to drifting gratings, grouped by genotype. (F) Distribution of lifetime sparseness in response to a drifting grating stimulus, grouped by genotype. In panels (C–F), colored numbers indicate the Jensen–Shannon distance between the wild-type distribution and the distributions of the four GCaMP-expressing mouse lines. [Siegle et al., 2021]#
However, we do observe some differences in behavior across the different transgenic lines. Most notably, we find that the locomotor behavior and eye movements of mice vary across different transgenic lines. [de Vries et al., 2020] Comparing the mean running speed across mice from different transgenic lines, we find significant differences. This largely reflects differences in the amount to time the mice spend running - rather than the mice all running the same amount but at different speeds. Likewise, we see differences in the number of saccades made by mice across transgenic lines. The latter appear to potentially be related to the reporter line more than the driver lines, though this has not been confirmed.
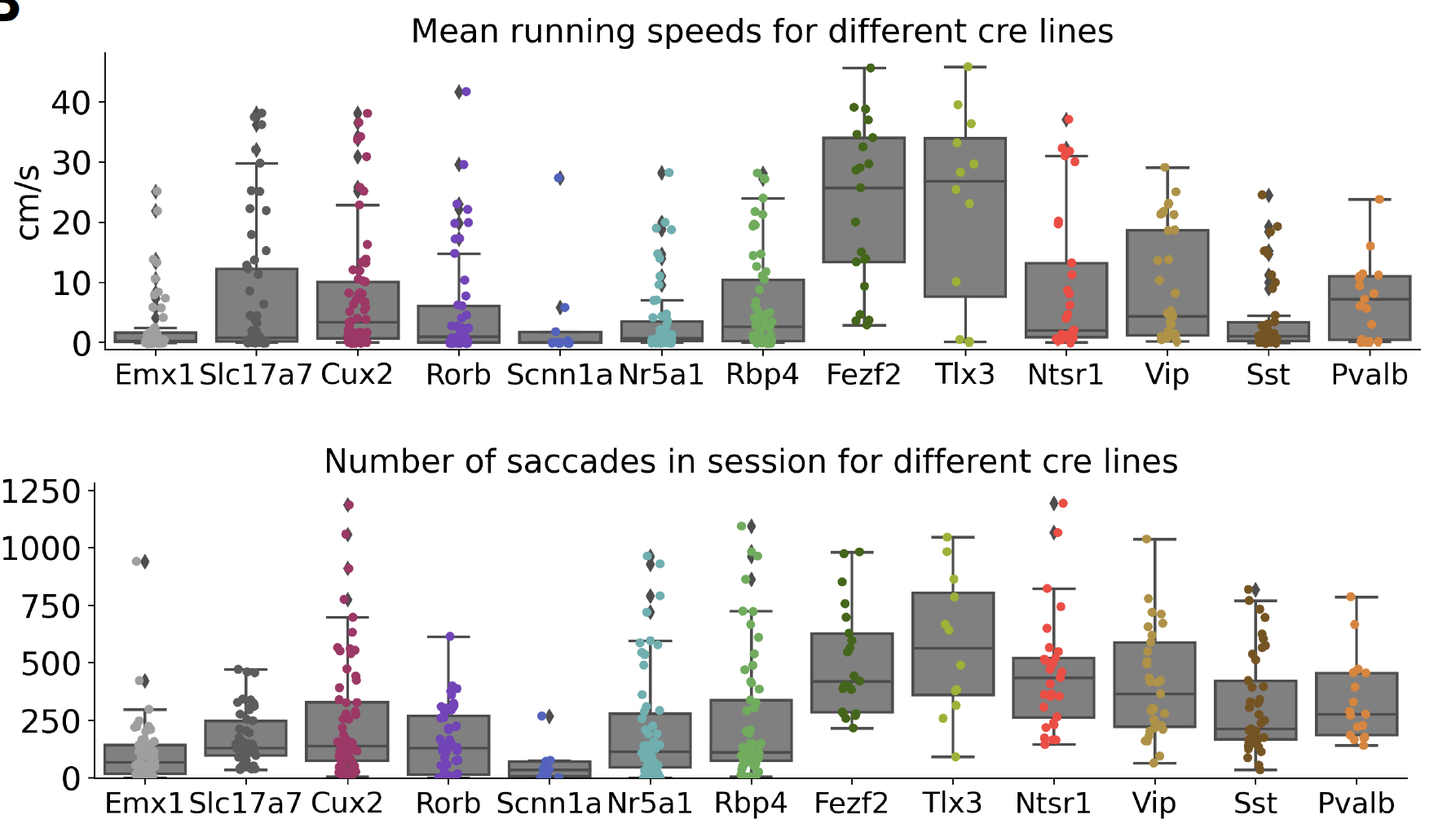
Fig. 5 (Top) Distribution of mean running speeds per imaging session for the Visual Coding 2P dataset, per transgenic line. Each dot is a single session. (Bottom) Distribution of number of saccades made per imaging session for the Visual Coding 2P dataset, per transgenic line. Each dot is a single session.#
