Electron Microscopy#
What Datasets are available for this course#
This year we have two cubic millimeter EM datasets available to work with: MICrONS and V1DD
The MICrONS Dataset#
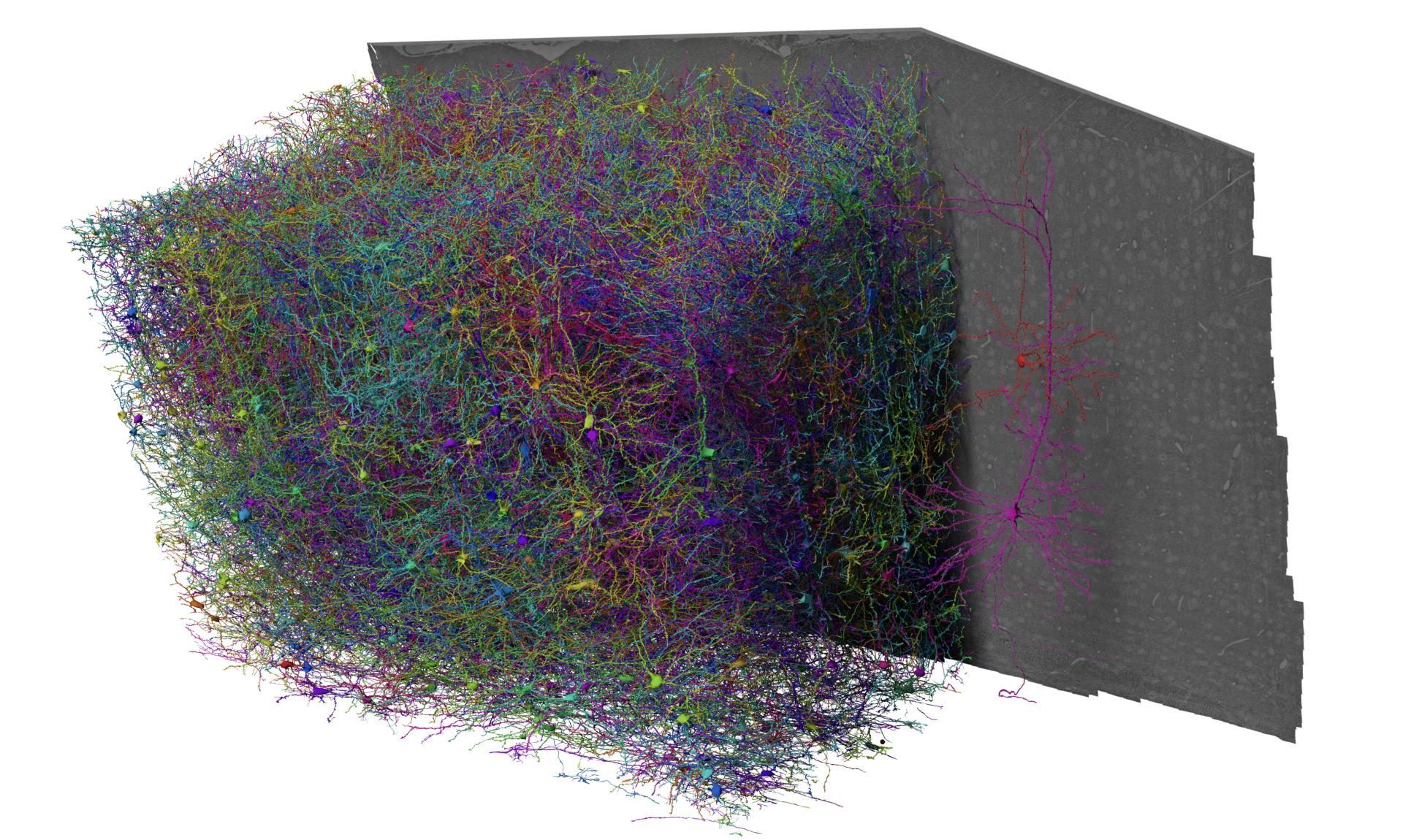
Fig. 24 The MICrONS Dataset, collected from 2018-2020, publicly available since 2021 but formally released April 2025#
The MICrONs Cubic Millimeter dataset is an EM volume of mouse visual cortex that spans all cortical layers, and extends approximately 1 mm in width by 0.5 mm in depth, collected as a collaboration between the Allen Institute for Brain Science, Baylor College of Medicine, and Princeton University. The goal of this dataset was to produce a so-called “functional connectomics” volume, where the same population of neurons could be measured with both calcium imaging to record functional properties and EM to describe synapse-resolution structural properties.
To link structure and function, excitatory neurons were genetically labeled with GCaMPs and imaged under a battery of natural movies and parametric stimuli by the Tolias lab, then at Baylor College of Medicine. The mouse was shipped to the Allen Institute and the same cortical region was prepared for EM, with more than 25,000 sections cut from the block and imaged across a small fleet of electron microscopes. In order to capture the interactions between different visual regions, the volume location is at the edge of primary visual cortex (VISp) and extends into higher visual areas VISal and VISrl.
Imagery was then aligned and segmented by the team of Sebastian Seung at Princeton, who also automatically detected synapses and nuclei. A team of proofreaders at Johns Hopkins Applied Physics Laboratory helped correct the largest types of errors in the segmentation, leaving virtually every neuron as a distinct object, and coregister tens of thousands of neurons to the functional data. Further analysis at the Allen Institute and elsewhere has focused on curating a map of cell types across the entire dataset, and generating a database to link synapses, cell types, and more to the morphology of individual cells.
The result is a cubic millimeter scale dataset with a rich collection of information and the potential for a range of studies, with strengths and limitations due to the current state of proofreading and needs of the functional data collection.
The V1 Deep-Dive (V1DD)#

Fig. 25 The V1 Deep-Dive (V1DD) Dataset, collected from 2020-2022, publicly available since August 2025#
Characterizing the relationship between neural function and connectivity is a central problem in visual sensory processing. In order to explore this relationship, we have recorded visual responses from pan-excitatory neurons within an 800X800 um region of primary visual cortex, spanning all visual layers from pia to white matter. This includes ~50,000 neurons per mouse in 4 mice total, collected from 150 2-photon and 7 3-photon calcium imaging planes spaced by ~16 um. This dataset will be used to examine the single-cell and population activity in primary visual cortex, and along with electron microscopic reconstruction from the same tissue, will serve as a valuable resource in studying the functional connectome in mouse cortex. A wide variety of visual stimuli were used to characterize neural responses, including windowed and full field drifting gratings, locally sparse noise, natural movies and natural images. We assessed multiple metrics, including receptive field profile, direction and orientation selectivity indices, reliability of response, and sparseness of response.
See more about the V1DD Physiology under the Physiology section of the databook.
These data are poised to focus on questions of visual encoding and population coding. Of particular focus is questions regarding surround suppression, where having a stimulus that extends beyond a neurons classical receptive field can suppress its response. Comparing the responses to the windowed and full field gratings can reveal such surround suppression - which can potentially be related to differences in connectivity patterns. Surround suppression has also been found to be stronger in superficial layers (e.g. layer 2/3) compared to deeper layers (e.g. layer 5), which can be explored in this dataset.
Other visual stimuli are also valuable for exploring the arrangement of spatial receptive fields or the correlations of neurons within a volume in response to natural movies or spontaneous activity. The behavior variables, including running speed/duration, and pupil area/position, are also of interest.
