Compare Trial Types#
The following example shows how to access behavioral and neural data for a given recording session and create plots for different trial types
Make sure that you have the AllenSDK installed in your environment
Imports#
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib notebook
%matplotlib inline
# Import allenSDK and check the version, which should be >2.10.2
import allensdk
allensdk.__version__
'2.16.2'
# import the behavior ophys project cache class from SDK to be able to load the data
from allensdk.brain_observatory.behavior.behavior_project_cache import VisualBehaviorOphysProjectCache
/opt/envs/allensdk/lib/python3.10/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
Load the cache and get data for one experiment#
# Set the path to the dataset
cache_dir = '/root/capsule/data/'
# If you are working with data in the cloud in Code Ocean,
# or if you have already downloaded the full dataset to your local machine,
# you can instantiate a local cache
# cache = VisualBehaviorOphysProjectCache.from_local_cache(cache_dir=cache_dir, use_static_cache=True)
# If you are working with the data locally for the first time, you need to instantiate the cache from S3:
cache = VisualBehaviorOphysProjectCache.from_local_cache(cache_dir=cache_dir, use_static_cache=True)
#cache = VisualBehaviorOphysProjectCache.from_s3_cache(cache_dir=cache_dir)
/opt/envs/allensdk/lib/python3.10/site-packages/allensdk/brain_observatory/behavior/behavior_project_cache/behavior_project_cache.py:135: UpdatedStimulusPresentationTableWarning:
As of AllenSDK version 2.16.0, the latest Visual Behavior Ophys data has been significantly updated from previous releases. Specifically the user will need to update all processing of the stimulus_presentations tables. These tables now include multiple stimulus types delineated by the columns `stimulus_block` and `stimulus_block_name`.
The data that was available in previous releases are stored in the block name containing 'change_detection' and can be accessed in the pandas table by using:
`stimulus_presentations[stimulus_presentations.stimulus_block_name.str.contains('change_detection')]`
warnings.warn(
ophys_experiment_table = cache.get_ophys_experiment_table()
Look at a sample of the experiment table#
ophys_experiment_table.sample(5)
| behavior_session_id | ophys_session_id | ophys_container_id | mouse_id | indicator | full_genotype | driver_line | cre_line | reporter_line | sex | ... | passive | experience_level | prior_exposures_to_session_type | prior_exposures_to_image_set | prior_exposures_to_omissions | date_of_acquisition | equipment_name | published_at | isi_experiment_id | file_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ophys_experiment_id | |||||||||||||||||||||
| 992084627 | 991992557 | 991958444 | 1018027778 | 485152 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | M | ... | False | Familiar | 0 | 28 | 0 | 2019-12-03 11:47:46.103000+00:00 | MESO.1 | 2021-03-25 | 949762677 | 1077 |
| 977247476 | 976558967 | 976382032 | 1018028215 | 482853 | GCaMP6f | Sst-IRES-Cre/wt;Ai148(TIT2L-GC6f-ICL-tTA2)/wt | [Sst-IRES-Cre] | Sst-IRES-Cre | Ai148(TIT2L-GC6f-ICL-tTA2) | M | ... | True | Familiar | 0 | 27 | 2 | 2019-11-05 08:53:47.241000+00:00 | MESO.1 | 2021-03-25 | 937755139 | 1465 |
| 969841037 | 969623770 | 969477369 | 1018028021 | 477052 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | M | ... | False | Novel >1 | 0 | 3 | 6 | 2019-10-17 14:59:42.873000+00:00 | MESO.1 | 2021-08-12 | 922677831 | 621 |
| 1089128033 | 1089052619 | 1089037437 | 1086191447 | 550603 | GCaMP6f | Vip-IRES-Cre/wt;Ai148(TIT2L-GC6f-ICL-tTA2)/wt | [Vip-IRES-Cre] | Vip-IRES-Cre | Ai148(TIT2L-GC6f-ICL-tTA2) | F | ... | False | Novel 1 | 0 | 0 | 6 | 2021-03-09 12:58:32.070000+00:00 | MESO.1 | 2021-08-12 | 1064781085 | 1757 |
| 939471248 | 938991494 | 938898514 | 1018028396 | 453989 | GCaMP6f | Vip-IRES-Cre/wt;Ai148(TIT2L-GC6f-ICL-tTA2)/wt | [Vip-IRES-Cre] | Vip-IRES-Cre | Ai148(TIT2L-GC6f-ICL-tTA2) | M | ... | True | Familiar | 0 | 52 | 1 | 2019-09-04 15:32:34.106000+00:00 | MESO.1 | 2021-03-25 | 848540715 | 1847 |
5 rows × 30 columns
Here are all of the unique session types#
np.sort(ophys_experiment_table['session_type'].unique())
array(['OPHYS_1_images_A', 'OPHYS_1_images_B', 'OPHYS_1_images_G',
'OPHYS_2_images_A_passive', 'OPHYS_2_images_B_passive',
'OPHYS_2_images_G_passive', 'OPHYS_3_images_A', 'OPHYS_3_images_B',
'OPHYS_3_images_G', 'OPHYS_4_images_A', 'OPHYS_4_images_B',
'OPHYS_4_images_H', 'OPHYS_5_images_A_passive',
'OPHYS_5_images_B_passive', 'OPHYS_5_images_H_passive',
'OPHYS_6_images_A', 'OPHYS_6_images_B', 'OPHYS_6_images_H'],
dtype=object)
Select an OPHYS_1_images_A experiment, load the experiment data#
experiment_id = ophys_experiment_table.query('session_type == "OPHYS_1_images_A"').sample(random_state=10).index[0]
print('getting experiment data for experiment_id {}'.format(experiment_id))
ophys_experiment = cache.get_behavior_ophys_experiment(experiment_id)
getting experiment data for experiment_id 1085840400
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
Look at task performance data#
We can see that the d-prime metric, a measure of discrimination performance, peaked at 2.14 during this session, indicating mid-range performance.
(d’ = 0 means no discrimination performance, d’ is infinite for perfect performance, but is limited to about 4.5 this dataset due to trial count limitations).
ophys_experiment.get_performance_metrics()
{'trial_count': 454,
'go_trial_count': 334,
'catch_trial_count': 48,
'hit_trial_count': 33,
'miss_trial_count': 301,
'false_alarm_trial_count': 1,
'correct_reject_trial_count': 47,
'auto_reward_count': 5,
'earned_reward_count': 33,
'total_reward_count': 38,
'total_reward_volume': 0.256,
'maximum_reward_rate': 2.860105184605357,
'engaged_trial_count': 68,
'mean_hit_rate': 0.12196894640281876,
'mean_hit_rate_uncorrected': 0.1148300747554362,
'mean_hit_rate_engaged': 0.2722870777110328,
'mean_false_alarm_rate': 0.07059889644968179,
'mean_false_alarm_rate_uncorrected': 0.020957227763510482,
'mean_false_alarm_rate_engaged': 0.05736544779097969,
'mean_dprime': 0.019241657181077048,
'mean_dprime_engaged': 0.9736703202887197,
'max_dprime': 1.321681073934908,
'max_dprime_engaged': 1.3119564166604674}
We can build a trial dataframe that tells us about behavior events on every trial. This can be merged with a rolling performance dataframe, which calculates behavioral performance metrics over a rolling window of 100 trials (excluding aborted trials, or trials where the animal licks prematurely).
trials_df = ophys_experiment.trials.merge(
ophys_experiment.get_rolling_performance_df().fillna(method='ffill'), # performance data is NaN on aborted trials. Fill forward to populate.
left_index = True,
right_index = True
)
trials_df.head()
| start_time | stop_time | initial_image_name | change_image_name | is_change | change_time | go | catch | lick_times | response_time | ... | aborted | auto_rewarded | change_frame | trial_length | reward_rate | hit_rate_raw | hit_rate | false_alarm_rate_raw | false_alarm_rate | rolling_dprime | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| trials_id | |||||||||||||||||||||
| 0 | 308.98272 | 316.25530 | im065 | im077 | True | 312.021593 | False | False | [312.48555, 312.73578, 312.90259, 313.05271, 3... | 312.48555 | ... | False | True | 18166 | 7.27258 | NaN | NaN | NaN | NaN | NaN | NaN |
| 1 | 316.48885 | 316.85579 | im077 | im077 | False | NaN | False | False | [316.5389] | NaN | ... | True | False | -99 | 0.36694 | NaN | NaN | NaN | NaN | NaN | NaN |
| 2 | 317.23947 | 318.17356 | im077 | im077 | False | NaN | False | False | [317.87332] | NaN | ... | True | False | -99 | 0.93409 | NaN | NaN | NaN | NaN | NaN | NaN |
| 3 | 318.74067 | 328.26515 | im077 | im061 | True | 324.031413 | False | False | [324.42868, 324.56211, 324.69556, 324.84573, 3... | 324.42868 | ... | False | True | 18886 | 9.52448 | NaN | NaN | NaN | NaN | NaN | NaN |
| 4 | 328.49870 | 337.30587 | im061 | im077 | True | 333.055463 | False | False | [333.48607, 333.73628, 333.90307, 334.05318, 3... | 333.48607 | ... | False | True | 19427 | 8.80717 | NaN | NaN | NaN | NaN | NaN | NaN |
5 rows × 27 columns
Now we can plot performance over the full experiment duration#
fig, ax = plt.subplots(2, 1, figsize = (15,5), sharex=True)
ax[0].plot(trials_df['start_time']/60., trials_df['hit_rate'], color='darkgreen')
ax[0].plot(trials_df['start_time']/60., trials_df['false_alarm_rate'], color='darkred')
ax[0].legend(['rolling hit rate', 'rolling false alarm rate'])
ax[1].plot(trials_df['start_time']/60., trials_df['rolling_dprime'], color='black')
ax[1].set_xlabel('trial start time (minutes)')
ax[0].set_ylabel('response rate')
ax[0].set_title('hit and false alarm rates')
ax[1].set_title("d'")
fig.tight_layout()
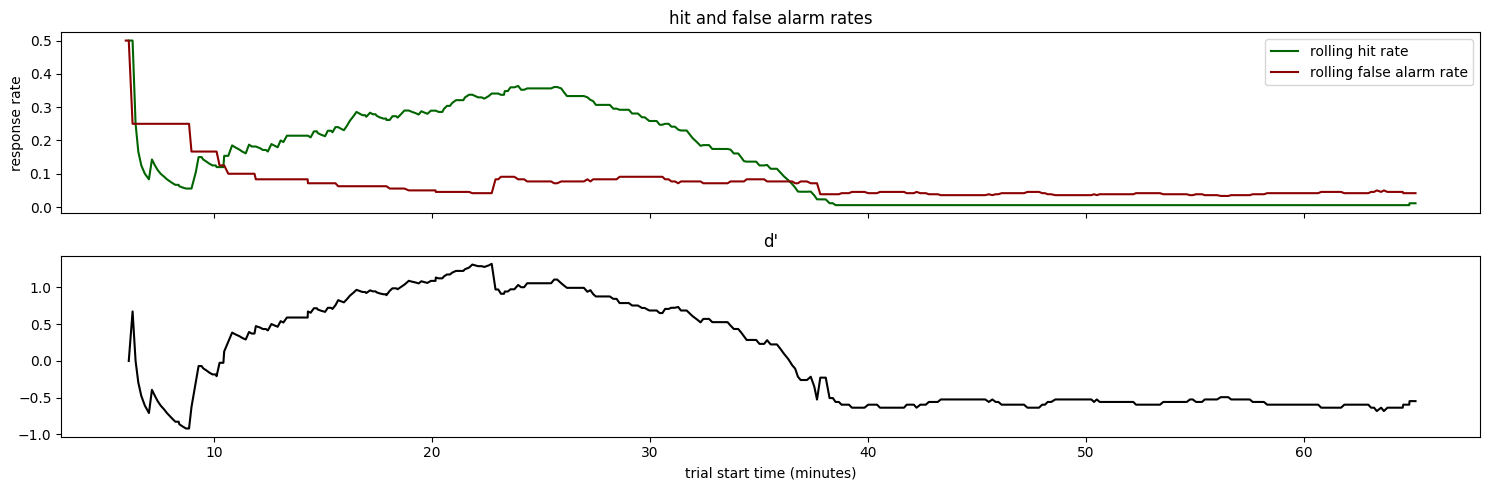
Some key observations:
The hit rate remains high for the first ~46 minutes of the session
The false alarm rate gradual declines during the first ~25 minutes of the session.
d’ peaks when the hit rate is still high, but the false alarm rate dips
The hit rate and d’ fall off dramatically after ~46 minutes. This is likely due to the animal becoming sated and losing motivation to perform
Plot neural data, behavior, and stimulus information for a trial#
Stimulus presentations#
Lets look at the dataframe of stimulus presentations. This tells us the attributes of every stimulus that was shown in the session
stimulus_presentations = ophys_experiment.stimulus_presentations
stimulus_presentations.head()
| stimulus_block | stimulus_block_name | image_index | image_name | movie_frame_index | duration | start_time | end_time | start_frame | end_frame | is_change | is_image_novel | omitted | movie_repeat | flashes_since_change | trials_id | is_sham_change | stimulus_name | active | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| stimulus_presentations_id | |||||||||||||||||||
| 0 | 0 | initial_gray_screen_5min | -99 | NaN | -99 | 309.019143 | 0.000000 | 309.019143 | 0 | 17986 | False | <NA> | <NA> | -99 | 0 | -99 | False | spontaneous | False |
| 1 | 1 | change_detection_behavior | 0 | im065 | -99 | 0.250200 | 309.019143 | 309.269343 | 17986 | 18001 | False | False | False | -99 | 1 | 0 | False | Natural_Images_Lum_Matched_set_training_2017 | True |
| 2 | 1 | change_detection_behavior | 0 | im065 | -99 | 0.250180 | 309.769753 | 310.019933 | 18031 | 18046 | False | False | False | -99 | 2 | 0 | False | Natural_Images_Lum_Matched_set_training_2017 | True |
| 3 | 1 | change_detection_behavior | 0 | im065 | -99 | 0.250200 | 310.520343 | 310.770543 | 18076 | 18091 | False | False | False | -99 | 3 | 0 | False | Natural_Images_Lum_Matched_set_training_2017 | True |
| 4 | 1 | change_detection_behavior | 0 | im065 | -99 | 0.250200 | 311.270953 | 311.521153 | 18121 | 18136 | False | False | False | -99 | 4 | 0 | False | Natural_Images_Lum_Matched_set_training_2017 | True |
To select information about stimuli in change detection behavior task only, we need to filter our table by stimulus block name.
stimulus_presentations = stimulus_presentations[stimulus_presentations.stimulus_block_name=='change_detection_behavior'].copy()
Note that there is an image name called ‘omitted’. This represents the time that a stimulus would have been shown, had it not been omitted from the regular stimulus cadence. They are included here for ease of analysis, but it’s important to note that they are not actually stimuli. They are the lack of expected stimuli.
stimulus_presentations.query('image_name == "omitted"').head()
| stimulus_block | stimulus_block_name | image_index | image_name | movie_frame_index | duration | start_time | end_time | start_frame | end_frame | is_change | is_image_novel | omitted | movie_repeat | flashes_since_change | trials_id | is_sham_change | stimulus_name | active | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| stimulus_presentations_id | |||||||||||||||||||
| 17 | 1 | change_detection_behavior | 8 | omitted | -99 | 0.25 | 321.028963 | 321.278963 | 18706 | 18721 | False | <NA> | True | -99 | 11 | 3 | False | Natural_Images_Lum_Matched_set_training_2017 | True |
| 25 | 1 | change_detection_behavior | 8 | omitted | -99 | 0.25 | 327.033833 | 327.283833 | 19066 | 19081 | False | <NA> | True | -99 | 3 | 3 | False | Natural_Images_Lum_Matched_set_training_2017 | True |
| 37 | 1 | change_detection_behavior | 8 | omitted | -99 | 0.25 | 336.057873 | 336.307873 | 19607 | 19622 | False | <NA> | True | -99 | 3 | 4 | False | Natural_Images_Lum_Matched_set_training_2017 | True |
| 63 | 1 | change_detection_behavior | 8 | omitted | -99 | 0.25 | 355.573823 | 355.823823 | 20777 | 20792 | False | <NA> | True | -99 | 4 | 7 | False | Natural_Images_Lum_Matched_set_training_2017 | True |
| 66 | 1 | change_detection_behavior | 8 | omitted | -99 | 0.25 | 357.825693 | 358.075693 | 20912 | 20927 | False | <NA> | True | -99 | 6 | 8 | False | Natural_Images_Lum_Matched_set_training_2017 | True |
Running speed#
One entry for each read of the analog input line monitoring the encoder voltage, polled at ~60 Hz.
ophys_experiment.running_speed.head()
| timestamps | speed | |
|---|---|---|
| 0 | 8.97085 | -0.027868 |
| 1 | 8.98751 | 0.056828 |
| 2 | 9.00421 | 0.121953 |
| 3 | 9.02091 | 0.154047 |
| 4 | 9.03756 | 0.148629 |
Licks#
One entry for every detected lick onset time, assigned the time of the corresponding visual stimulus frame.
ophys_experiment.licks.head()
| timestamps | frame | |
|---|---|---|
| 0 | 10.88909 | 115 |
| 1 | 13.17429 | 252 |
| 2 | 13.25767 | 257 |
| 3 | 13.35777 | 263 |
| 4 | 13.47453 | 270 |
Eye tracking data#
One entry containing ellipse fit parameters for the eye, pupil and corneal reflection for every frame of the eye tracking video stream.
ophys_experiment.eye_tracking.head()
| timestamps | cr_area | eye_area | pupil_area | likely_blink | pupil_area_raw | cr_area_raw | eye_area_raw | cr_center_x | cr_center_y | ... | eye_center_x | eye_center_y | eye_width | eye_height | eye_phi | pupil_center_x | pupil_center_y | pupil_width | pupil_height | pupil_phi | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| frame | |||||||||||||||||||||
| 0 | 0.12895 | 120.778985 | 52646.046976 | 9467.868055 | False | 9467.868055 | 120.778985 | 52646.046976 | 304.855877 | 238.850452 | ... | 321.614711 | 229.150072 | 148.455382 | 112.880766 | -0.040543 | 284.593634 | 219.583658 | 54.897322 | 46.068901 | -0.741087 |
| 1 | 0.15369 | 115.944914 | 52448.649649 | 10145.403510 | False | 10145.403510 | 115.944914 | 52448.649649 | 304.125448 | 240.159601 | ... | 320.796911 | 229.198209 | 148.349421 | 112.537842 | -0.036200 | 282.597206 | 216.890346 | 56.827654 | 43.674891 | -0.650310 |
| 2 | 0.17215 | 120.194858 | 52110.324268 | 10430.167825 | False | 10430.167825 | 120.194858 | 52110.324268 | 304.436750 | 242.135068 | ... | 319.853374 | 231.950317 | 148.468978 | 111.721867 | -0.046359 | 283.692110 | 223.351216 | 47.981925 | 57.619663 | 0.643652 |
| 3 | 0.21069 | 112.516058 | 51693.903652 | 10825.284266 | False | 10825.284266 | 112.516058 | 51693.903652 | 304.122429 | 241.159250 | ... | 321.063479 | 230.763062 | 148.371626 | 110.901802 | -0.042321 | 282.785915 | 222.177822 | 46.984507 | 58.700894 | 0.606463 |
| 4 | 0.24880 | 112.314878 | 52186.596867 | 10278.561636 | False | 10278.561636 | 112.314878 | 52186.596867 | 304.529129 | 239.723128 | ... | 321.497389 | 228.294415 | 148.304450 | 112.009517 | -0.038966 | 283.189635 | 219.170357 | 47.385991 | 57.199369 | 0.654353 |
5 rows × 23 columns
Neural data as deltaF/F#
One row per cell, with each containing an array of deltaF/F values.
ophys_experiment.dff_traces.head()
| cell_roi_id | dff | |
|---|---|---|
| cell_specimen_id | ||
| 1120118128 | 1115339979 | [2.7728134118137593, 2.7377723001149357, 1.332... |
| 1120118208 | 1115339983 | [0.3466408785876189, 0.4530561213911281, 0.347... |
| 1120118626 | 1115340014 | [0.6086096210226783, 0.65493126738475, 0.73593... |
| 1120118698 | 1115340017 | [0.9173731088455804, 0.282220484734575, 0.3718... |
| 1120118967 | 1115340030 | [0.30710398969593, 0.44648909591684693, 0.0910... |
We can convert the dff_traces to long-form (aka “tidy”) as follows:#
def get_cell_timeseries_dict(dataset, cell_specimen_id):
'''
for a given cell_specimen ID, this function creates a dictionary with the following keys
* timestamps: ophys timestamps
* cell_roi_id
* cell_specimen_id
* dff
This is useful for generating a tidy dataframe, which can enable easier plotting of timeseries data
arguments:
session object
cell_specimen_id
returns
dict
'''
cell_dict = {
'timestamps': dataset.ophys_timestamps,
'cell_roi_id': [dataset.dff_traces.loc[cell_specimen_id]['cell_roi_id']] * len(dataset.ophys_timestamps),
'cell_specimen_id': [cell_specimen_id] * len(dataset.ophys_timestamps),
'dff': dataset.dff_traces.loc[cell_specimen_id]['dff'],
}
return cell_dict
ophys_experiment.tidy_dff_traces = pd.concat(
[pd.DataFrame(get_cell_timeseries_dict(ophys_experiment, cell_specimen_id)) for cell_specimen_id in ophys_experiment.dff_traces.reset_index()['cell_specimen_id']]).reset_index(drop=True)
ophys_experiment.tidy_dff_traces.sample(5)
| timestamps | cell_roi_id | cell_specimen_id | dff | |
|---|---|---|---|---|
| 430740 | 4239.62011 | 1115340053 | 1120134575 | 0.089869 |
| 6711575 | 1266.16980 | 1115340580 | 1120133562 | 0.186147 |
| 4572603 | 4024.04204 | 1115340449 | 1120132573 | -0.126489 |
| 5962321 | 3300.71198 | 1115340537 | 1120133228 | 0.005886 |
| 5288889 | 3412.94295 | 1115340491 | 1120132857 | -0.183917 |
Plot all the data streams for different trial types#
We can look at a few trial types in some detail
Define plotting functions#
First define functions to plot the different data streams:
each stimulus as a colored vertical bar
running speed
licks/rewards
pupil area
neural responses (dF/F)
def add_image_colors(stimulus_presentations):
'''
Add a column to stimulus_presentations called 'color' with a unique color for each image in the session
'''
# gather image names but exclude image_name=='omitted'
unique_stimuli = [stimulus for stimulus in stimulus_presentations['image_name'].unique() if stimulus != 'omitted']
# assign a color for each unique stimulus
colormap = {image_name: sns.color_palette()[image_number] for image_number, image_name in enumerate(np.sort(unique_stimuli))}
colormap['omitted'] = [1, 1, 1] # assign white to omitted
# add color column to stimulus presentations
stimulus_presentations['color'] = stimulus_presentations['image_name'].map(lambda image_name: colormap[image_name])
return stimulus_presentations
def plot_stimuli(trial, ax):
'''
plot stimuli as colored bars on specified axis
'''
stimuli = ophys_experiment.stimulus_presentations.query("stimulus_block_name == 'change_detection_behavior'").copy()
stimuli = add_image_colors(stimuli)
stimuli = stimuli[(stimuli.end_time >= trial['start_time'].values[0]) &
(stimuli.start_time <= trial['stop_time'].values[0])]
for idx, stimulus in stimuli.iterrows():
ax.axvspan(stimulus['start_time'], stimulus['end_time'], color=stimulus['color'], alpha=0.5)
return ax
def plot_running(trial, ax):
'''
plot running speed for trial on specified axes
'''
trial_running_speed = ophys_experiment.running_speed.copy()
trial_running_speed = trial_running_speed[(trial_running_speed.timestamps >= trial['start_time'].values[0]) &
(trial_running_speed.timestamps <= trial['stop_time'].values[0])]
ax.plot(trial_running_speed['timestamps'], trial_running_speed['speed'], color='black')
ax.set_title('running speed')
ax.set_ylabel('speed (cm/s)')
return ax
def plot_licks(trial, ax):
'''
plot licks as black dots on specified axis
'''
trial_licks = ophys_experiment.licks.copy()
trial_licks = trial_licks[(trial_licks.timestamps >= trial['start_time'].values[0]) &
(trial_licks.timestamps <= trial['stop_time'].values[0])]
ax.plot(trial_licks['timestamps'], np.zeros_like(trial_licks['timestamps']),
marker = 'o', linestyle = 'none', color='black')
return ax
def plot_rewards(trial, ax):
'''
plot rewards as blue diamonds on specified axis
'''
trial_rewards = ophys_experiment.rewards.copy()
trial_rewards = trial_rewards[(trial_rewards.timestamps >= trial['start_time'].values[0]) &
(trial_rewards.timestamps <= trial['stop_time'].values[0])]
ax.plot(trial_rewards['timestamps'], np.zeros_like(trial_rewards['timestamps']),
marker = 'd', linestyle = 'none', color='blue', markersize = 10, alpha = 0.25)
return ax
def plot_pupil(trial, ax):
'''
plot pupil area on specified axis
'''
trial_eye_tracking = ophys_experiment.eye_tracking.copy()
trial_eye_tracking = trial_eye_tracking[(trial_eye_tracking.timestamps >= trial['start_time'].values[0]) &
(trial_eye_tracking.timestamps <= trial['stop_time'].values[0])]
ax.plot(trial_eye_tracking['timestamps'], trial_eye_tracking['pupil_area'], color='black')
ax.set_title('pupil area')
ax.set_ylabel('pupil area\n')
return ax
def plot_dff(trial, ax):
'''
plot each cell's dff response for a given trial
'''
# get the tidy dataframe of dff traces we created earlier
trial_dff_traces = ophys_experiment.tidy_dff_traces.copy()
# filter to get this trial
trial_dff_traces = trial_dff_traces[(trial_dff_traces.timestamps >= trial['start_time'].values[0]) &
(trial_dff_traces.timestamps <= trial['stop_time'].values[0])]
# plot each cell
for cell_specimen_id in ophys_experiment.tidy_dff_traces['cell_specimen_id'].unique():
ax.plot(trial_dff_traces[trial_dff_traces.cell_specimen_id == cell_specimen_id]['timestamps'],
trial_dff_traces[trial_dff_traces.cell_specimen_id == cell_specimen_id]['dff'])
ax.set_title('deltaF/F responses')
ax.set_ylabel('dF/F')
return ax
def make_trial_plot(trial):
'''
combine all plots for a given trial
'''
fig, axes = plt.subplots(4, 1, figsize = (15, 8), sharex=True)
for ax in axes:
plot_stimuli(trial, ax)
plot_running(trial, axes[0])
plot_licks(trial, axes[1])
plot_rewards(trial, axes[1])
axes[1].set_title('licks and rewards')
axes[1].set_yticks([])
axes[1].legend(['licks','rewards'])
plot_pupil(trial, axes[2])
plot_dff(trial, axes[3])
axes[3].set_xlabel('time in session (seconds)')
fig.tight_layout()
return fig, axes
Here is a hit trial#
stimulus_presentations.columns
Index(['stimulus_block', 'stimulus_block_name', 'image_index', 'image_name',
'movie_frame_index', 'duration', 'start_time', 'end_time',
'start_frame', 'end_frame', 'is_change', 'is_image_novel', 'omitted',
'movie_repeat', 'flashes_since_change', 'trials_id', 'is_sham_change',
'stimulus_name', 'active'],
dtype='object')
trials = ophys_experiment.trials.copy()
trial = trials[trials.hit==True].sample()
fig, axes = make_trial_plot(trial)
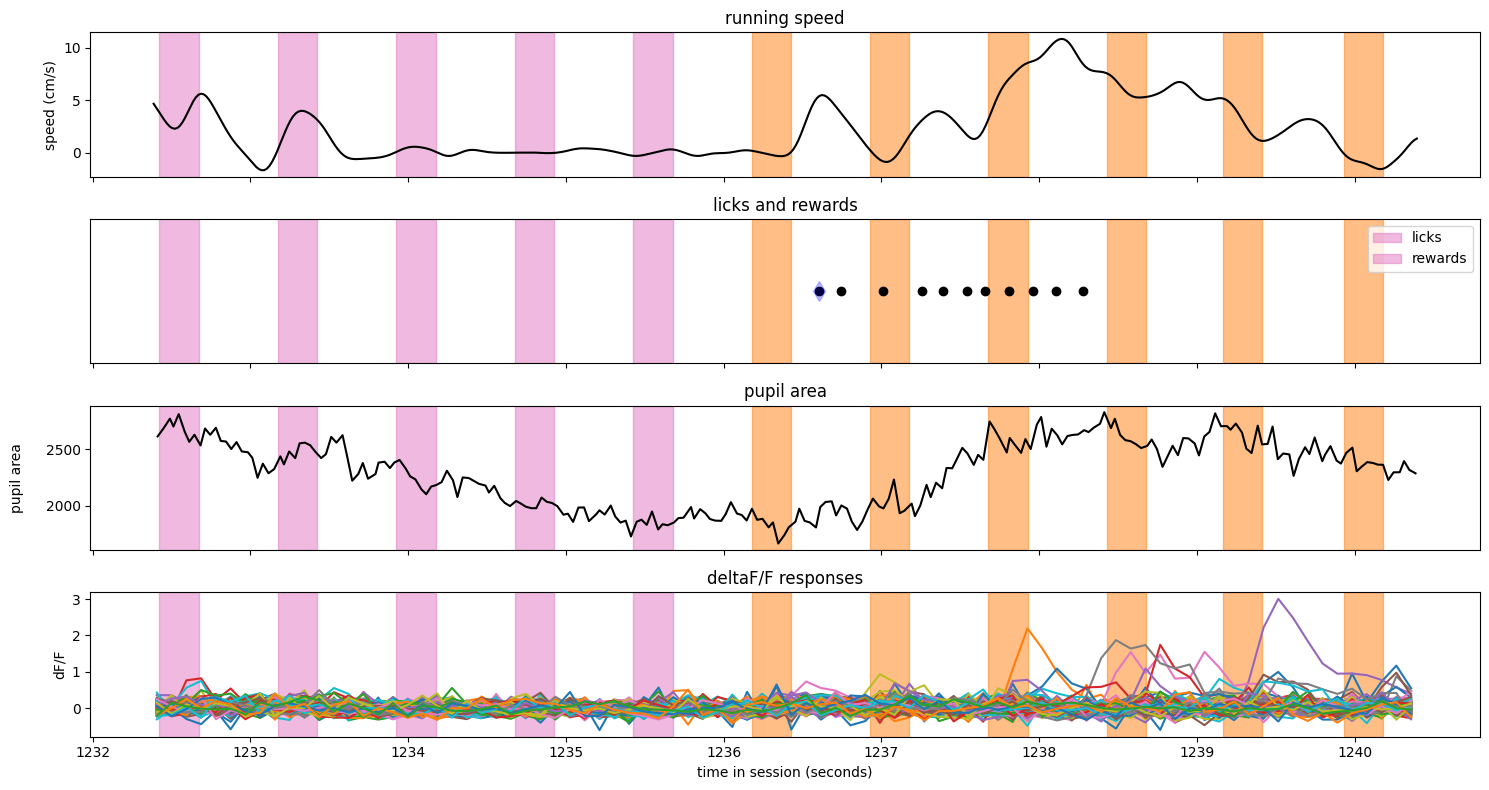
Notes:
The image identity changed just after t = 2361 seconds (note the color change in the vertical spans)
The animal was running steadily prior to the image change, then slowed to a stop after the change
The first lick occurred about 500 ms after the change, and triggered an immediate reward
The pupil area shows some missing data - these were points that were filtered out as outliers.
There appears to be one neuron that was responding regularly to the stimulus prior to the change.
Here is a miss trial#
trial = ophys_experiment.trials.query('miss').sample()
fig, axes = make_trial_plot(trial)
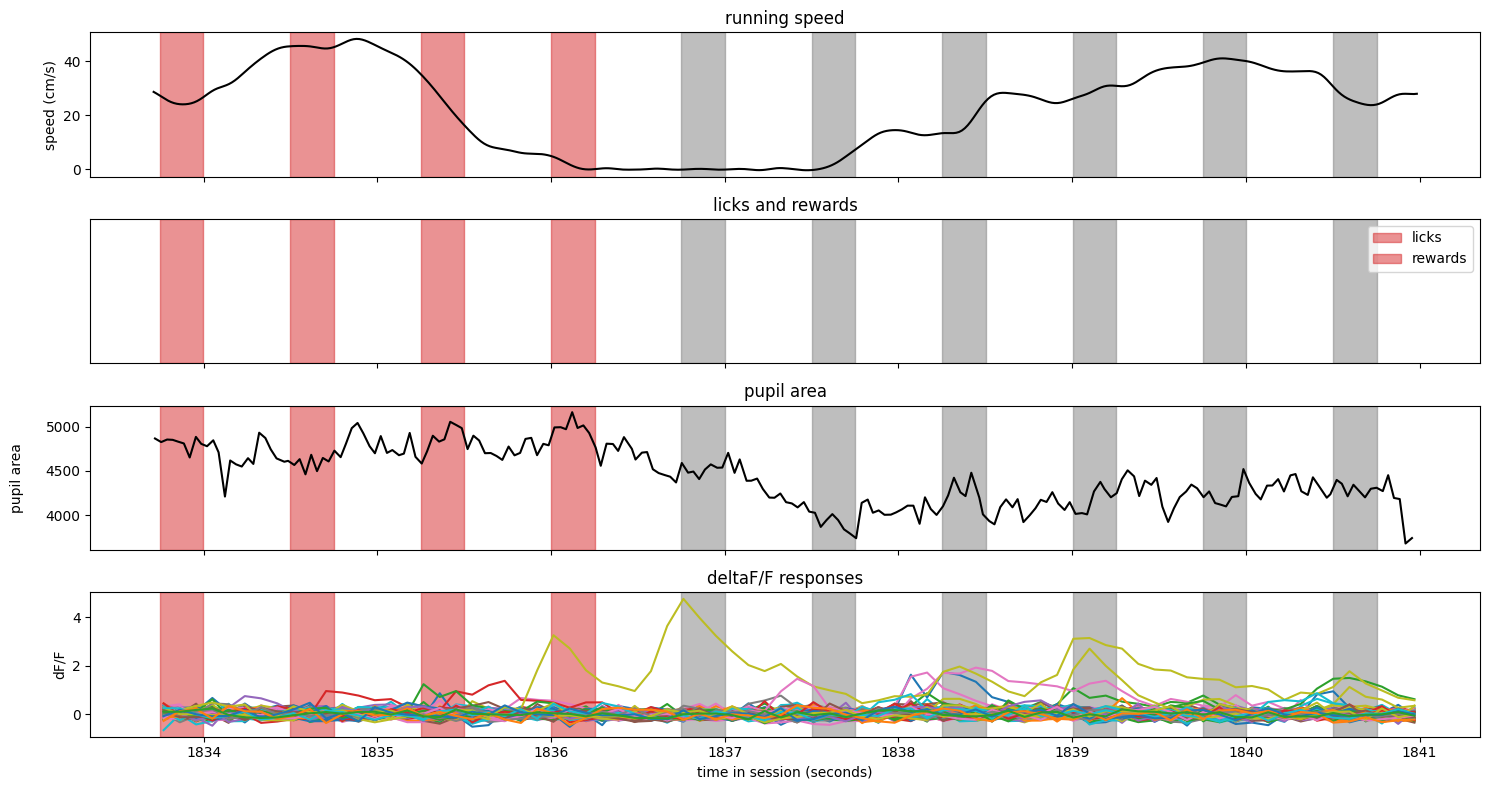
Notes:
The image identity changed just after t = 824 seconds (note the color change in the vertical spans)
The animal was running relatively steadily during the entire trial and did not slow after the stimulus identity change
There were no licks or rewards on this trial
The pupil area shows some missing data - these were points that were filtered out as outliers.
One neuron had a large response just prior to the change, but none appear to be stimulus locked on this trial
Here is a false alarm trial#
trial = ophys_experiment.trials.query('false_alarm').sample()
fig, axes = make_trial_plot(trial)
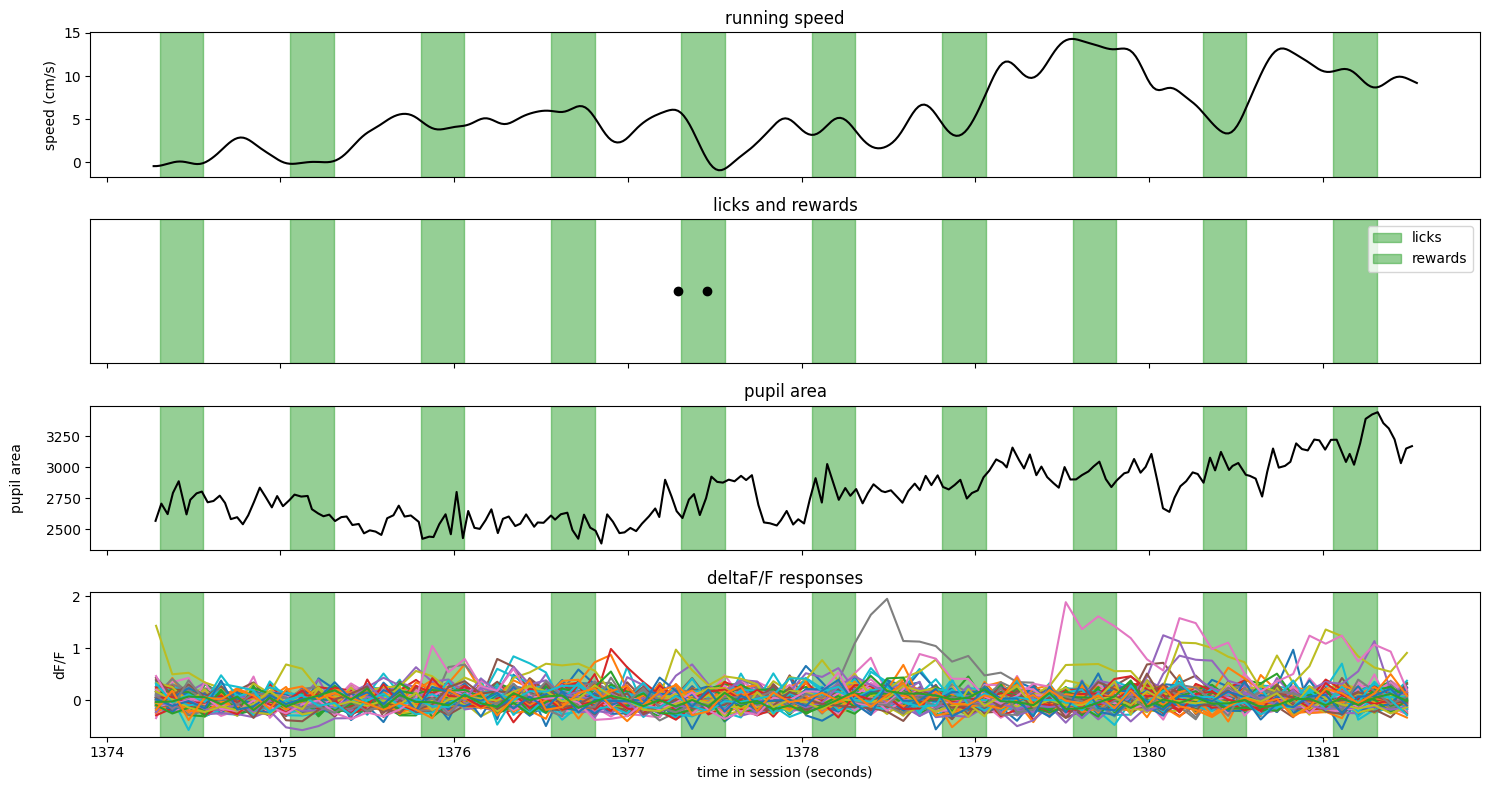
Notes:
The image identity was consistent during the entire trial
The animal slowed and licked partway through the trial
There were no rewards on this trial
The pupil area shows some missing data - these were points that were filtered out as outliers.
There were not any neurons with obvious stimulus locked responses
And finally, a correct rejection#
trial = ophys_experiment.trials.query('correct_reject').sample()
fig, axes = make_trial_plot(trial)
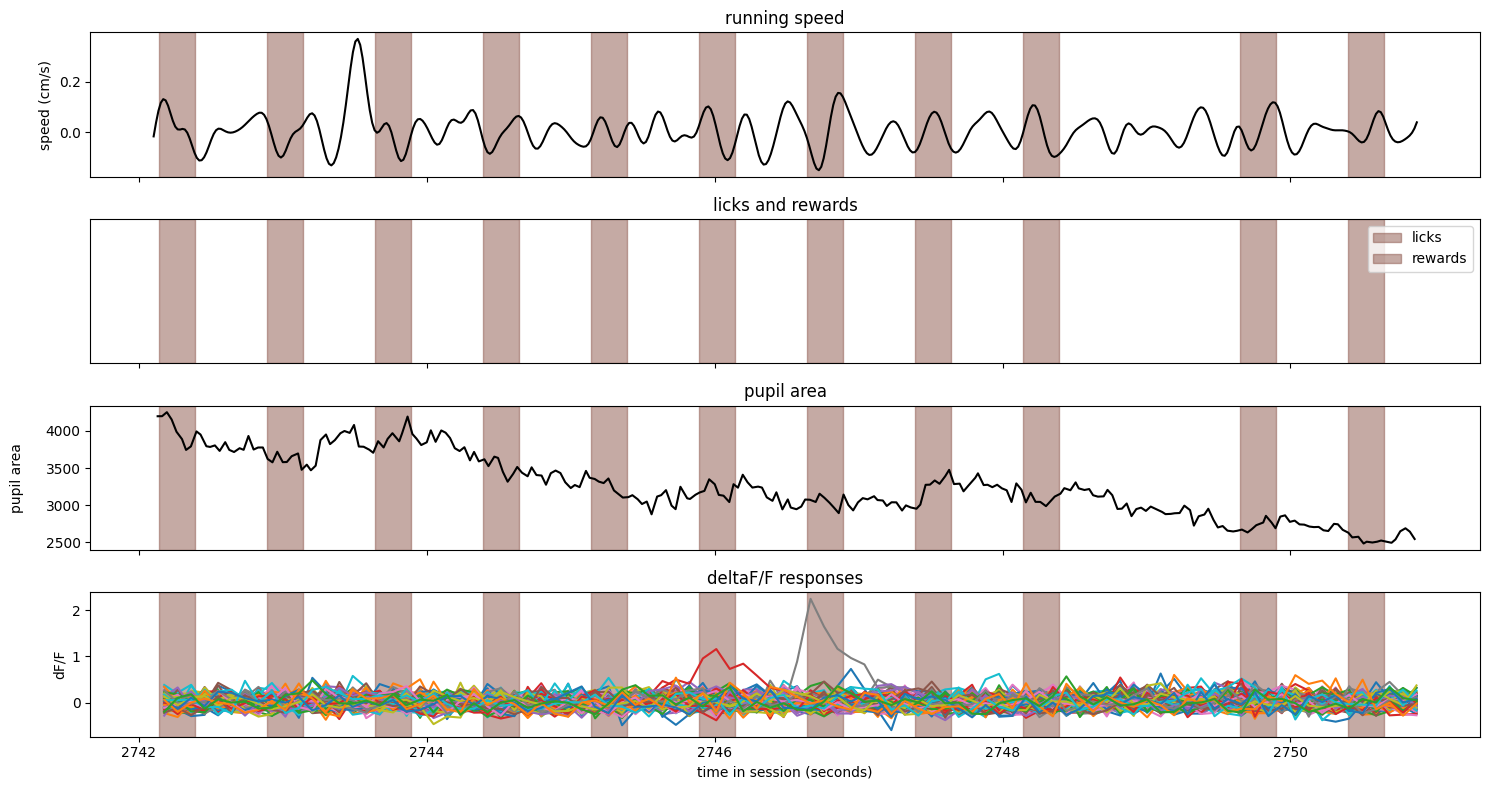
Notes:
The image identity was consistent during the entire trial
The animal did not slow or lick during this trial
There were no rewards on this trial
