Visual Behavior Neuropixels Quickstart#
A short introduction to the Visual Behavior Neuropixels data and SDK. This notebook focuses on aligning neural data to visual and optotagging stimuli. To learn more about how to access the data, see our data access tutorial. For more information about task and behavioral data, check out the other tutorials accompanying this dataset.
Also note that this project shares many features with the Visual Coding Neuropixels and Visual Behavior 2-Photon datasets. Users are encouraged to check out the documentation for those projects for additional information and context.
Contents#
import os
from pathlib import Path
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from allensdk.brain_observatory.behavior.behavior_project_cache.\
behavior_neuropixels_project_cache \
import VisualBehaviorNeuropixelsProjectCache
%matplotlib inline
/opt/envs/allensdk/lib/python3.10/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
The VisualBehaviorNeuropixelsProjectCache is the main entry point to the Visual Behavior Neuropixels dataset. It allows you to download data for individual recording sessions and view cross-session summary information.
# Update this to a valid directory in your filesystem. This is where the data will be stored.
cache_dir = '/root/capsule/data/'
cache = VisualBehaviorNeuropixelsProjectCache.from_local_cache(
cache_dir=cache_dir, use_static_cache=True)
# get the metadata tables
units_table = cache.get_unit_table()
channels_table = cache.get_channel_table()
probes_table = cache.get_probe_table()
behavior_sessions_table = cache.get_behavior_session_table()
ecephys_sessions_table = cache.get_ecephys_session_table()
This dataset contains ephys recording sessions from 3 genotypes (C57BL6J, VIP-IRES-CrexAi32 and SST-IRES-CrexAi32). For each mouse, two recordings were made on consecutive days. One of these sessions used the image set that was familiar to the mouse from training. The other session used a novel image set containing two familiar images from training and six new images that the mouse had never seen. As an example, let’s grab a session from an SST mouse during a novel session.
sst_novel_sessions = ecephys_sessions_table.loc[(ecephys_sessions_table['genotype'].str.contains('Sst')) &
(ecephys_sessions_table['experience_level']=='Novel')]
sst_novel_sessions.head()
| behavior_session_id | date_of_acquisition | equipment_name | session_type | mouse_id | genotype | sex | project_code | age_in_days | unit_count | ... | channel_count | structure_acronyms | image_set | prior_exposures_to_image_set | session_number | experience_level | prior_exposures_to_omissions | file_id | abnormal_histology | abnormal_activity | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ecephys_session_id | |||||||||||||||||||||
| 1048189115 | 1048221709 | 2020-09-03 14:16:57.913000+00:00 | NP.1 | EPHYS_1_images_H_3uL_reward | 509808 | Sst-IRES-Cre/wt;Ai32(RCL-ChR2(H134R)_EYFP)/wt | M | NeuropixelVisualBehavior | 264 | 1925 | ... | 2304 | ['APN', 'CA1', 'CA3', 'DG-mo', 'DG-po', 'DG-sg... | H | 0 | 2 | Novel | 1 | 879 | NaN | NaN |
| 1048196054 | 1048222325 | 2020-09-03 14:25:07.290000+00:00 | NP.0 | EPHYS_1_images_H_3uL_reward | 524925 | Sst-IRES-Cre/wt;Ai32(RCL-ChR2(H134R)_EYFP)/wt | F | NeuropixelVisualBehavior | 166 | 2288 | ... | 2304 | ['APN', 'CA1', 'CA3', 'DG-mo', 'DG-po', 'DG-sg... | H | 0 | 2 | Novel | 1 | 880 | NaN | NaN |
| 1053941483 | 1053960987 | 2020-10-01 17:03:58.362000+00:00 | NP.1 | EPHYS_1_images_H_3uL_reward | 527749 | Sst-IRES-Cre/wt;Ai32(RCL-ChR2(H134R)_EYFP)/wt | M | NeuropixelVisualBehavior | 180 | 1543 | ... | 2304 | ['APN', 'CA1', 'CA3', 'DG-mo', 'DG-po', 'DG-sg... | H | 0 | 2 | Novel | 1 | 891 | NaN | NaN |
| 1064644573 | 1064666428 | 2020-11-19 15:18:01.372000+00:00 | NP.1 | EPHYS_1_images_H_3uL_reward | 544456 | Sst-IRES-Cre/wt;Ai32(RCL-ChR2(H134R)_EYFP)/wt | M | NeuropixelVisualBehavior | 120 | 2254 | ... | 2304 | ['APN', 'CA1', 'CA3', 'DG-mo', 'DG-po', 'DG-sg... | H | 0 | 2 | Novel | 1 | 909 | NaN | NaN |
| 1065905010 | 1065929713 | 2020-11-24 14:21:48.847000+00:00 | NP.0 | EPHYS_1_images_H_3uL_reward | 544358 | Sst-IRES-Cre/wt;Ai32(RCL-ChR2(H134R)_EYFP)/wt | F | NeuropixelVisualBehavior | 126 | 1998 | ... | 2304 | ['APN', 'CA1', 'CA3', 'DG-mo', 'DG-po', 'DG-sg... | H | 0 | 2 | Novel | 1 | 912 | NaN | NaN |
5 rows × 21 columns
Now let’s choose one of these sessions to look at more closely.
session_id = 1064644573
session = cache.get_ecephys_session(
ecephys_session_id=session_id)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
We can get a high-level summary of this session by accessing its metadata attribute:
session.metadata
{'equipment_name': 'NP.1',
'sex': 'M',
'age_in_days': 120,
'stimulus_frame_rate': 60.0,
'session_type': 'EPHYS_1_images_H_3uL_reward',
'date_of_acquisition': datetime.datetime(2020, 11, 19, 23, 18, 1, tzinfo=tzutc()),
'reporter_line': 'Ai32(RCL-ChR2(H134R)_EYFP)',
'cre_line': 'Sst-IRES-Cre',
'behavior_session_uuid': UUID('731026fa-d60b-4819-be73-aa9d757c84ed'),
'driver_line': ['Sst-IRES-Cre'],
'mouse_id': '544456',
'project_code': 'Not Available',
'full_genotype': 'Sst-IRES-Cre/wt;Ai32(RCL-ChR2(H134R)_EYFP)/wt',
'behavior_session_id': 1064666428,
'ecephys_session_id': 1064644573}
Now we can get the unit and channel data for this session. Merging these dataframes will give us CCF coordinates for each unit
units = session.get_units()
channels = session.get_channels()
unit_channels = units.merge(channels, left_on='peak_channel_id', right_index=True)
Let’s look at which brain structures were recorded during this session
unit_channels.value_counts('structure_acronym')
structure_acronym
APN 304
CA1 295
DG 190
VISp 144
VISpm 134
VISl 118
VISal 106
MB 103
CA3 100
LP 99
VISam 96
VISrl 92
MGv 86
MRN 79
ProS 70
NB 56
PIL 56
POST 32
NOT 27
Eth 15
root 14
TH 11
SUB 9
HPF 9
MGd 7
POL 1
MGm 1
dtype: int64
PSTH for image changes#
Now we’ll grab spike times and calculate the change response for ‘good’ units in V1. Note that how you filter units will depend on your analysis. Consult the unit metrics notebook for more details.
#first let's sort our units by depth
unit_channels = unit_channels.sort_values('probe_vertical_position', ascending=False)
#now we'll filter them
good_unit_filter = ((unit_channels['snr']>1)&
(unit_channels['isi_violations']<1)&
(unit_channels['firing_rate']>0.1))
good_units = unit_channels.loc[good_unit_filter]
spike_times = session.spike_times
We can get the times when the image changes occurred from the stimulus presentations table. For now, we’ll only take the image changes shown during the active behavior block
stimulus_presentations = session.stimulus_presentations
change_times = stimulus_presentations[stimulus_presentations['active']&
stimulus_presentations['is_change']]['start_time'].values
#Convenience function to compute the PSTH
def makePSTH(spikes, startTimes, windowDur, binSize=0.001):
bins = np.arange(0,windowDur+binSize,binSize)
counts = np.zeros(bins.size-1)
for i,start in enumerate(startTimes):
startInd = np.searchsorted(spikes, start)
endInd = np.searchsorted(spikes, start+windowDur)
counts = counts + np.histogram(spikes[startInd:endInd]-start, bins)[0]
counts = counts/startTimes.size
return counts/binSize, bins
We’ll include enough time in our plot to see three image responses: the pre-change image response, the change response and the post-change response
#Here's where we loop through the units in our area of interest and compute their PSTHs
area_of_interest = 'VISp'
area_change_responses = []
area_units = good_units[good_units['structure_acronym']==area_of_interest]
time_before_change = 1
duration = 2.5
for iu, unit in area_units.iterrows():
unit_spike_times = spike_times[iu]
unit_change_response, bins = makePSTH(unit_spike_times,
change_times-time_before_change,
duration, binSize=0.01)
area_change_responses.append(unit_change_response)
area_change_responses = np.array(area_change_responses)
#Plot the results
fig, ax = plt.subplots(1,2)
fig.set_size_inches([12,4])
clims = [np.percentile(area_change_responses, p) for p in (0.1,99.9)]
im = ax[0].imshow(area_change_responses, clim=clims)
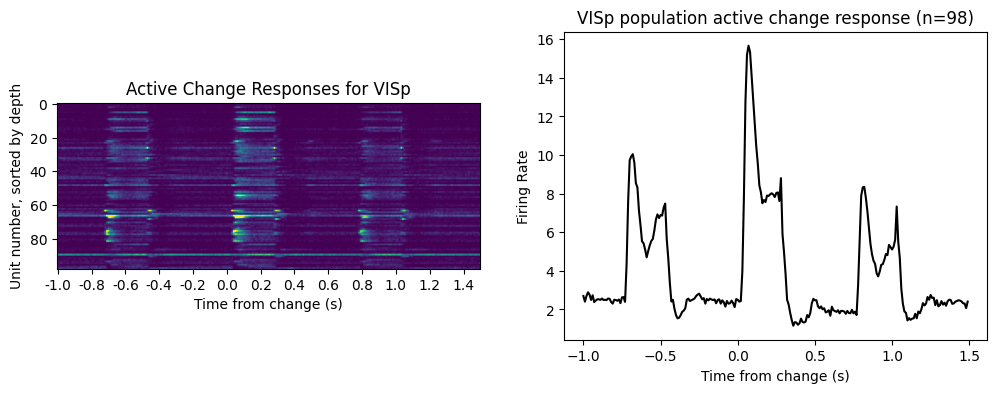
ax[0].set_title('Active Change Responses for {}'.format(area_of_interest))
ax[0].set_ylabel('Unit number, sorted by depth')
ax[0].set_xlabel('Time from change (s)')
ax[0].set_xticks(np.arange(0, bins.size-1, 20))
_ = ax[0].set_xticklabels(np.round(bins[:-1:20]-time_before_change, 2))
ax[1].plot(bins[:-1]-time_before_change, np.mean(area_change_responses, axis=0), 'k')
ax[1].set_title('{} population active change response (n={})'\
.format(area_of_interest, area_change_responses.shape[0]))
ax[1].set_xlabel('Time from change (s)')
ax[1].set_ylabel('Firing Rate')
Text(0, 0.5, 'Firing Rate')
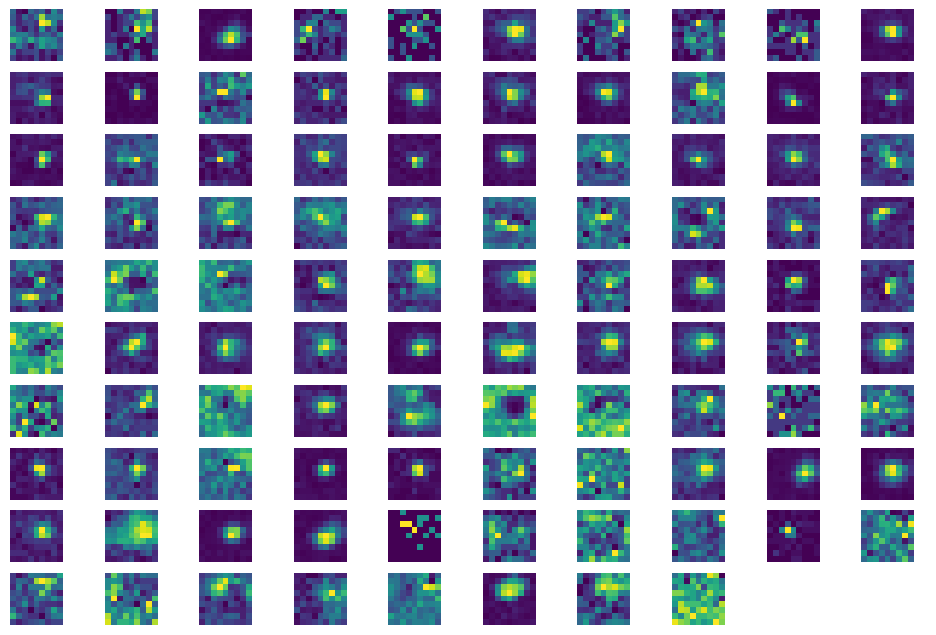
Plot Receptive Fields#
Now we’ll plot receptive fields for these same units. First we need to get stimulus presentation data for the receptive field mapping stimulus (gabors). For more information about this stimulus, consult this tutorial for the Visual Coding Neuropixels dataset (though note that not all the functionality in the visual coding SDK will work for this dataset).
rf_stim_table = stimulus_presentations[stimulus_presentations['stimulus_name'].str.contains('gabor')]
xs = np.sort(rf_stim_table.position_x.unique()) #positions of gabor along azimuth
ys = np.sort(rf_stim_table.position_y.unique()) #positions of gabor along elevation
def find_rf(spikes, xs, ys):
unit_rf = np.zeros([ys.size, xs.size])
for ix, x in enumerate(xs):
for iy, y in enumerate(ys):
stim_times = rf_stim_table[(rf_stim_table.position_x==x)
&(rf_stim_table.position_y==y)]['start_time'].values
unit_response, bins = makePSTH(spikes,
stim_times+0.01,
0.2, binSize=0.001)
unit_rf[iy, ix] = unit_response.mean()
return unit_rf
area_rfs = []
for iu, unit in area_units.iterrows():
unit_spike_times = spike_times[iu]
unit_rf = find_rf(unit_spike_times, xs, ys)
area_rfs.append(unit_rf)
fig, axes = plt.subplots(int(len(area_rfs)/10)+1, 10)
fig.set_size_inches(12, 8)
for irf, rf in enumerate(area_rfs):
ax_row = int(irf/10)
ax_col = irf%10
axes[ax_row][ax_col].imshow(rf, origin='lower')
for ax in axes.flat:
ax.axis('off')
Optotagging#
Since this is an SST mouse, we should see putative SST+ interneurons that are activated during our optotagging protocol. Let’s load the optotagging stimulus table and plot PSTHs triggered on the laser onset. For more examples and useful info about optotagging, you can check out the Visual Coding Neuropixels Optotagging notebook here (though note that not all the functionality in the visual coding SDK will work for this dataset).
opto_table = session.optotagging_table
opto_table.head()
| start_time | condition | level | stop_time | stimulus_name | duration | |
|---|---|---|---|---|---|---|
| id | ||||||
| 0 | 8801.01737 | half-period of a cosine wave | 0.950 | 8802.01737 | raised_cosine | 1.00 |
| 1 | 8803.02730 | a single square pulse | 0.800 | 8803.03730 | pulse | 0.01 |
| 2 | 8805.12189 | a single square pulse | 0.800 | 8805.13189 | pulse | 0.01 |
| 3 | 8806.91173 | a single square pulse | 0.800 | 8806.92173 | pulse | 0.01 |
| 4 | 8809.01283 | half-period of a cosine wave | 0.685 | 8810.01283 | raised_cosine | 1.00 |
If you check out this table, you’ll see that we use 2 different laser waveforms: a short square pulse that’s 10 ms long and a half-period cosine that’s 1 second long:
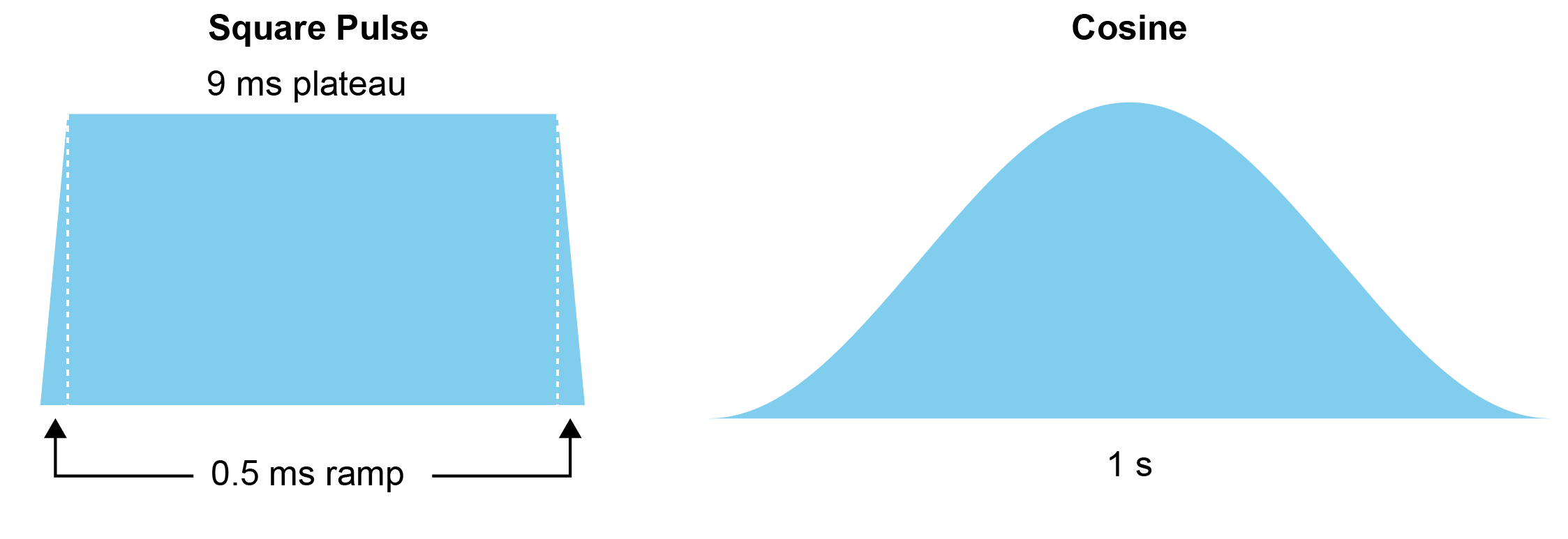
We drive each at three light levels, giving us 6 total conditions. Now let’s plot how cortical neurons respond to the short pulse at high power.
duration = opto_table.duration.min() #get the short pulses
level = opto_table.level.max() #and the high power trials
cortical_units = good_units[good_units['structure_acronym'].str.contains('VIS')]
opto_times = opto_table.loc[(opto_table['duration']==duration)&
(opto_table['level']==level)]['start_time'].values
time_before = 0.01 # seconds to take before the laser start for PSTH
duration = 0.03 # total duration of trial for PSTH in seconds
binSize = 0.001 # 1ms bin size for PSTH
opto_response = []
unit_id = []
for iu, unit in cortical_units.iterrows():
unit_spike_times = spike_times[iu]
unit_response, bins = makePSTH(unit_spike_times,
opto_times-time_before, duration,
binSize=binSize)
opto_response.append(unit_response)
unit_id.append(iu)
opto_response = np.array(opto_response)
fig, ax = plt.subplots()
fig.set_size_inches((5,10))
fig.suptitle('Optotagging: ' + str(session.metadata['ecephys_session_id'])
+ ' ' + session.metadata['full_genotype'])
im = ax.imshow(opto_response,
origin='lower', aspect='auto',
)
min_clim_val = 0
max_clim_val = 250
im.set_clim([min_clim_val, max_clim_val])
[ax.axvline(bound, linestyle=':', color='white', linewidth=1.0)\
for bound in [10, 19]]
ax.set_xlabel('Time from laser onset (ms)')
ax.set_ylabel('Unit number')
ax.set_xticks(1000*bins[:-1:5])
time_labels = np.round(1000*(bins[:-1:5]-time_before), 0)
_=ax.set_xticklabels(time_labels)
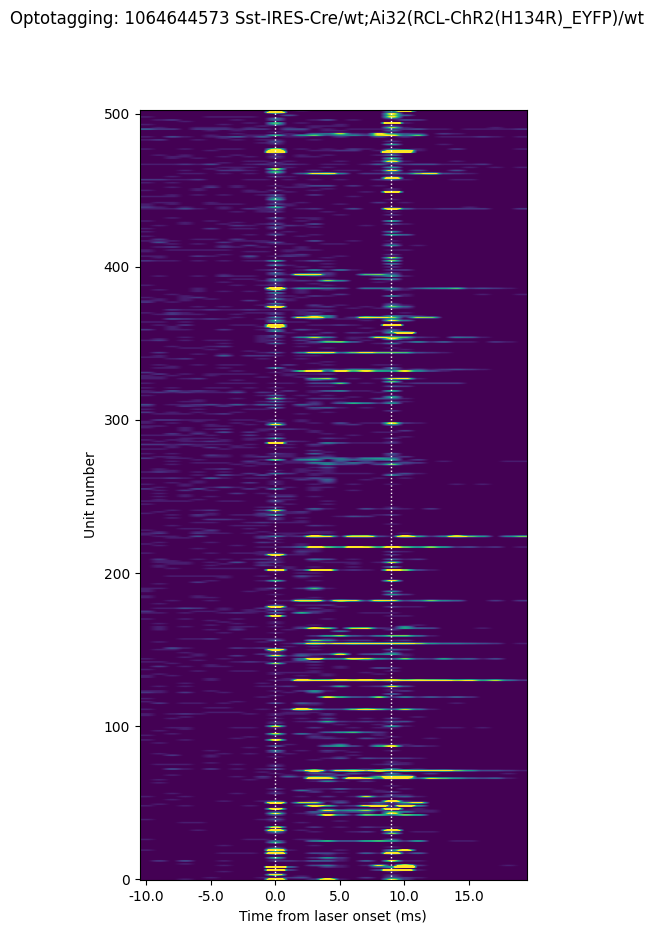
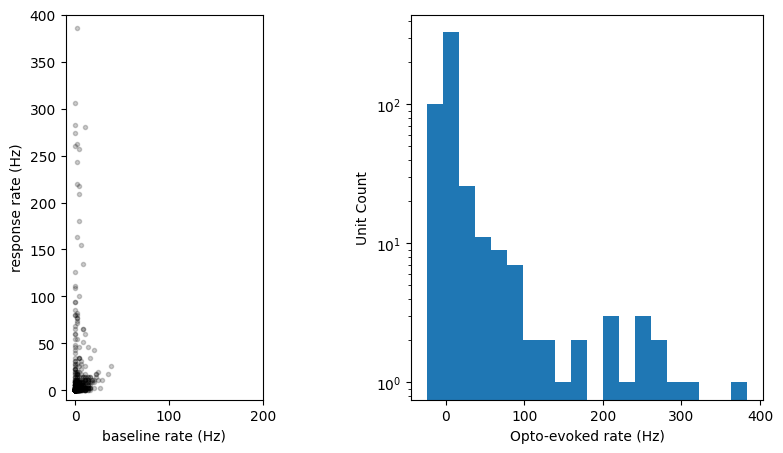
Here we can see that most units don’t respond to the short laser pulse. But there is a population that do show elevated firing rates. Note that the activity occurring at the onset and offset of the laser (along the dotted lines) is artifactual and should be excluded from analysis.
Let’s plot the response to the laser over the population:
baseline_window = slice(0, 9) # baseline epoch
response_window = slice(11,18) # laser epoch
response_magnitudes = np.mean(opto_response[:, response_window], axis=1) \
- np.mean(opto_response[:, baseline_window], axis=1)
fig, axes = plt.subplots(1,2)
fig.set_size_inches(10, 5)
# Plot scatter of opto rate vs baseline rate
axes[0].plot(np.mean(opto_response[:, baseline_window], axis=1),
np.mean(opto_response[:, response_window], axis=1), 'k.', alpha=0.2)
axes[0].set_xlim([-10, 200])
axes[0].set_ylim([-10, 400])
axes[0].set_aspect('equal')
axes[0].set_ylabel('response rate (Hz)')
axes[0].set_xlabel('baseline rate (Hz)')
# Plot histogram of opto-evoked rate (note log yscale)
_ = axes[1].hist(response_magnitudes, bins=20)
axes[1].set_yscale('log')
axes[1].set_xlabel('Opto-evoked rate (Hz)')
axes[1].set_ylabel('Unit Count')
Text(0, 0.5, 'Unit Count')