import pandas as pd
Dataset#
We will start exploring the parameters of the dataset to learn what data is available.
First we need to access the dataset. We will use the AllenSDK and the BrainObservatoryCache to do so. First we need to set this up - the key step is to provide a manifest file. The SDK uses this file to know what data is available and organize the files it downloads. If you instantiate the BrainObservatoryCache without providing a manifest file, it will create one in your working directory.
from allensdk.core.brain_observatory_cache import BrainObservatoryCache
manifest_file = '../../../data/allen-brain-observatory/visual-coding-2p/manifest.json'
boc = BrainObservatoryCache(manifest_file=manifest_file)
We can use the BrainObservatoryCache to explore the parameters of the dataset.
Targeted structures#
What brain regions were recorded across the dataset? To determine this we use a function called get_all_targeted_structures() to create a list of the regions.
boc.get_all_targeted_structures()
['VISal', 'VISam', 'VISl', 'VISp', 'VISpm', 'VISrl']
We see that data was collected in six different visual areas. VISp is the primary visual cortex, also known as V1. The others are higher visual areas (HVAs) that surround VISp. You can learn more about these areas and how we map them here.
Cre lines and reporters#
We used Cre lines to drive the expression of GCaMP6 in specific populations of neurons. We can find a list of all the cre lines used in this dataset with a similar function
boc.get_all_cre_lines()
['Cux2-CreERT2',
'Emx1-IRES-Cre',
'Fezf2-CreER',
'Nr5a1-Cre',
'Ntsr1-Cre_GN220',
'Pvalb-IRES-Cre',
'Rbp4-Cre_KL100',
'Rorb-IRES2-Cre',
'Scnn1a-Tg3-Cre',
'Slc17a7-IRES2-Cre',
'Sst-IRES-Cre',
'Tlx3-Cre_PL56',
'Vip-IRES-Cre']
Cre is a driver that drives the expression of a reporter. We used four different reporter lines in this dataset.
boc.get_all_reporter_lines()
['Ai148(TIT2L-GC6f-ICL-tTA2)',
'Ai162(TIT2L-GC6s-ICL-tTA2)',
'Ai93(TITL-GCaMP6f)',
'Ai93(TITL-GCaMP6f)-hyg',
'Ai94(TITL-GCaMP6s)']
Note
Reporter lines: All the experiments in this dataset use GCaMP6. The large majority use GCaMP6f and only a few use GCaMP6s. However, you see four different reporters listed here. Why is this? Ai93 is the GCaMP6f reporter we used with the excitatory Cre lines. However, this reporter does not work well for inhibitory Cre lines. We used Ai148, another GCaMP6f reporter, with Vip-IRES-Cre and Sst-IRES-Cre. However, this didn’t work with the Pvalb-IRES-Cre. We use Ai162, a GCaMP6s reporter with Pvalb. Additionally, to have a GCaMP6f vs GCaMP6s comparison, we collected a small number of experiments using Ai94 with the Slc17a7-IRES2-Cre. This is a GCaMP6s reporter that complements Ai93. Slc17a7-IRES2-Cre is the only Cre line that was recorded using multiple reporter types.
See Transgenic tools to learn more about these Cre lines and reporters.
Imaging depths#
Each experiment was collected at a single imaging depth.
boc.get_all_imaging_depths()
[175,
185,
195,
200,
205,
225,
250,
265,
275,
276,
285,
300,
320,
325,
335,
350,
365,
375,
390,
400,
550,
570,
625]
These values are in µm below the surface of the cortex. This is a long list and some of the values don’t differ by very much. How meaningful is it? We roughly consider depths less than 250 to be layer 2/3, 250-350 to be layer 4, 350-500 to be layer 5, and over 500 to be layer 6. Keep in mind, much of the imaging here was done with layer specific Cre lines, so for most purposes the best way to get layer specificity is to select appropriate Cre lines.
Visual stimuli#
What were the visual stimuli that we showed to the mice?
boc.get_all_stimuli()
['drifting_gratings',
'locally_sparse_noise',
'locally_sparse_noise_4deg',
'locally_sparse_noise_8deg',
'natural_movie_one',
'natural_movie_three',
'natural_movie_two',
'natural_scenes',
'spontaneous',
'static_gratings']
These are described more extensively in Visual stimuli.
Experiment containers & sessions#
The experiment container describes a set of 3 imaging sessions performed for the same field of view (ie. same targeted structure and imaging depth in the same mouse that targets the same set of neurons). Each experiment container has a unique ID number.
We will identify all the experiment containers for a given structure and Cre line:
visual_area = 'VISp'
cre_line ='Cux2-CreERT2'
exps = boc.get_experiment_containers(targeted_structures=[visual_area], cre_lines=[cre_line])
Note
get_experiment_containers returns all experiment containers that meet the conditions we have specified. The parameters that we could pass this function include targeted_structures, imaging_depths, cre_lines, reporter_lines, stimuli, session_types, and cell_specimen_ids. If we don’t pass any parameters, it returns all experiment containers.
We can make a dataframe of the list of experiment containers to see what information we get about them:
pd.DataFrame(exps)
| id | imaging_depth | targeted_structure | cre_line | reporter_line | donor_name | specimen_name | tags | failed | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 511510736 | 175 | VISp | Cux2-CreERT2 | Ai93(TITL-GCaMP6f) | 222426 | Cux2-CreERT2;Camk2a-tTA;Ai93-222426 | [] | False |
| 1 | 511510855 | 175 | VISp | Cux2-CreERT2 | Ai93(TITL-GCaMP6f) | 229106 | Cux2-CreERT2;Camk2a-tTA;Ai93-229106 | [] | False |
| 2 | 511509529 | 175 | VISp | Cux2-CreERT2 | Ai93(TITL-GCaMP6f) | 222420 | Cux2-CreERT2;Camk2a-tTA;Ai93-222420 | [] | False |
| 3 | 511507650 | 175 | VISp | Cux2-CreERT2 | Ai93(TITL-GCaMP6f) | 222424 | Cux2-CreERT2;Camk2a-tTA;Ai93-222424 | [] | False |
| 4 | 702934962 | 275 | VISp | Cux2-CreERT2 | Ai93(TITL-GCaMP6f) | 382421 | Cux2-CreERT2;Camk2a-tTA;Ai93-382421 | [] | False |
| 5 | 645413757 | 275 | VISp | Cux2-CreERT2 | Ai93(TITL-GCaMP6f) | 348262 | Cux2-CreERT2;Camk2a-tTA;Ai93-348262 | [] | False |
| 6 | 659767480 | 275 | VISp | Cux2-CreERT2 | Ai93(TITL-GCaMP6f) | 360565 | Cux2-CreERT2;Camk2a-tTA;Ai93-360565 | [] | False |
| 7 | 511510650 | 175 | VISp | Cux2-CreERT2 | Ai93(TITL-GCaMP6f) | 222425 | Cux2-CreERT2;Camk2a-tTA;Ai93-222425 | [] | False |
| 8 | 712178509 | 275 | VISp | Cux2-CreERT2 | Ai93(TITL-GCaMP6f) | 390323 | Cux2-CreERT2;Camk2a-tTA;Ai93-390323 | [] | False |
| 9 | 511510667 | 275 | VISp | Cux2-CreERT2 | Ai93(TITL-GCaMP6f) | 222420 | Cux2-CreERT2;Camk2a-tTA;Ai93-222420 | [] | False |
| 10 | 524691282 | 275 | VISp | Cux2-CreERT2 | Ai93(TITL-GCaMP6f) | 243293 | Cux2-CreERT2;Camk2a-tTA;Ai93-243293 | [] | False |
| 11 | 701412138 | 175 | VISp | Cux2-CreERT2 | Ai93(TITL-GCaMP6f) | 382421 | Cux2-CreERT2;Camk2a-tTA;Ai93-382421 | [] | False |
| 12 | 511510718 | 175 | VISp | Cux2-CreERT2 | Ai93(TITL-GCaMP6f) | 231584 | Cux2-CreERT2;Camk2a-tTA;Ai93-231584 | [] | False |
| 13 | 511510699 | 275 | VISp | Cux2-CreERT2 | Ai93(TITL-GCaMP6f) | 225037 | Cux2-CreERT2;Camk2a-tTA;Ai93-225037 | [] | False |
| 14 | 511510779 | 275 | VISp | Cux2-CreERT2 | Ai93(TITL-GCaMP6f) | 225036 | Cux2-CreERT2;Camk2a-tTA;Ai93-225036 | [] | False |
| 15 | 511510670 | 175 | VISp | Cux2-CreERT2 | Ai93(TITL-GCaMP6f) | 225037 | Cux2-CreERT2;Camk2a-tTA;Ai93-225037 | [] | False |
- id
The experiment container id
- imaging_depth
The imaging depth that data was acquired at, in um from the surface of cortex.
- targeted_structure
The brain structure that was imaged in this session.
- cre_line
The Cre line that the mouse has.
- reporter_line
The reporter line that the mouse has.
- donor_name
The id of the mouse that was imaged.
- specimen_name
The full name of the mouse including its genotype and donor name.
- tags
A list of tags
- failed
Boolean indicating whether the experiment container failed QC. By default, only container that pass QC are returned. Users must specify to include failed experiment containers if looking for these.
You see there are 16 experiments for this Cre line in VISp. They all have different experiment container ids (called “id” here) and they mostly have different donor names which identify the mouse that was imaged. This Cre line was imaged at two different imaging depths, sampling both layer 2/3 and layer 4. But they all have the same Cre line, targeted structure and reporter line.
Exercise: How many experiment containers were collected in each cortical visual area for each Cre line?#
cre_lines = boc.get_all_cre_lines()
areas = boc.get_all_targeted_structures()
df = pd.DataFrame(columns=areas, index=cre_lines)
for cre in cre_lines:
for area in areas:
exps = boc.get_experiment_containers(targeted_structures=[area], cre_lines=[cre])
df[area].loc[cre] = len(exps)
df
| VISal | VISam | VISl | VISp | VISpm | VISrl | |
|---|---|---|---|---|---|---|
| Cux2-CreERT2 | 13 | 11 | 11 | 16 | 13 | 12 |
| Emx1-IRES-Cre | 7 | 3 | 8 | 10 | 4 | 9 |
| Fezf2-CreER | 0 | 0 | 5 | 4 | 0 | 0 |
| Nr5a1-Cre | 6 | 6 | 6 | 8 | 7 | 6 |
| Ntsr1-Cre_GN220 | 0 | 0 | 7 | 6 | 5 | 0 |
| Pvalb-IRES-Cre | 0 | 0 | 5 | 16 | 0 | 0 |
| Rbp4-Cre_KL100 | 6 | 8 | 7 | 7 | 6 | 4 |
| Rorb-IRES2-Cre | 6 | 8 | 6 | 8 | 7 | 5 |
| Scnn1a-Tg3-Cre | 0 | 0 | 0 | 9 | 0 | 0 |
| Slc17a7-IRES2-Cre | 2 | 2 | 19 | 60 | 15 | 2 |
| Sst-IRES-Cre | 1 | 0 | 17 | 30 | 14 | 2 |
| Tlx3-Cre_PL56 | 0 | 0 | 3 | 6 | 0 | 0 |
| Vip-IRES-Cre | 0 | 0 | 24 | 36 | 16 | 0 |
You see that not all Cre lines were imaged in all areas.
Session types#
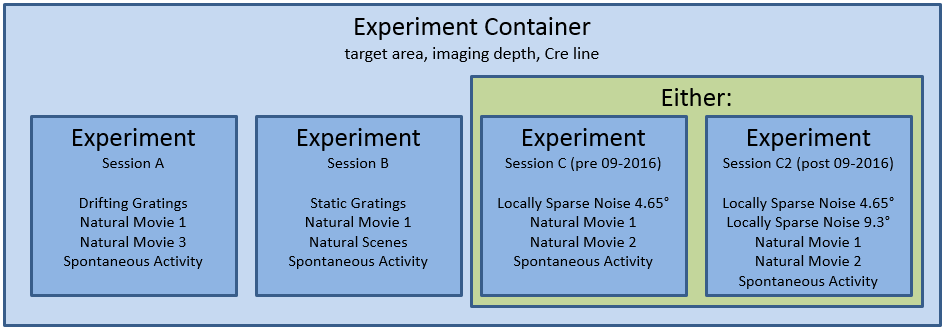
The responses to this full set of visual stimuli were recorded across three imaging sessions. We returned to the same targeted structure and same imaging depth in the same mouse to recorded the same group of neurons across three different days.
Let’s look at all of the sessions in a single experiment container.
experiment_container_id = 511510736
sessions = boc.get_ophys_experiments(experiment_container_ids=[experiment_container_id])
Note
Much like get_experiment_containers, get_ophys_experiments returns all experiment sessions that meet the conditions we have specified. The parameters that we could pass this function include targeted_structures, imaging_depths, cre_lines, reporter_lines, stimuli, session_types, experiment-container_id, and cell_specimen_ids. If we don’t pass any parameters, it returns all experiment sessions.
Let’s look at a DataFrame of the results
pd.DataFrame(sessions)
| id | imaging_depth | targeted_structure | cre_line | reporter_line | acquisition_age_days | experiment_container_id | session_type | donor_name | specimen_name | fail_eye_tracking | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 501559087 | 175 | VISp | Cux2-CreERT2 | Ai93(TITL-GCaMP6f) | 103 | 511510736 | three_session_B | 222426 | Cux2-CreERT2;Camk2a-tTA;Ai93-222426 | True |
| 1 | 501704220 | 175 | VISp | Cux2-CreERT2 | Ai93(TITL-GCaMP6f) | 104 | 511510736 | three_session_A | 222426 | Cux2-CreERT2;Camk2a-tTA;Ai93-222426 | True |
| 2 | 501474098 | 175 | VISp | Cux2-CreERT2 | Ai93(TITL-GCaMP6f) | 102 | 511510736 | three_session_C | 222426 | Cux2-CreERT2;Camk2a-tTA;Ai93-222426 | True |
- id
The session id for the session. This is the id that is used to access data for that session.
- imaging_depth
The imaging depth that data was acquired at, in um from the surface of cortex.
- targeted_structure
The brain structure that was imaged in this session.
- cre_line
The Cre line that the mouse has.
- reporter_line
The reporter line that the mouse has.
- acquisition_age_days
The age of the mouse when this session was recorded, in days.
- experiment_container_id
The id of the experiment container that this session belongs to.
- session_type
The name of the session, this describes the set of stimuli that are presented during the experiment.
- donor_name
The id of the mouse that was imaged.
- specimen_name
The full name of the mouse including its genotype and donor name.
- fail_eye_tracking
Boolean marking which sessions had successful eye tracking. This might be obsolete.
When looking at all of the sessions in a single experiment container, as we have done above, you will notice that the experiment container id, cre line, reporter line, donor name, specimen name, imaging depth, targeted structure are all the same while the id, acquisition age, and session type must be different.
As you see, each experiment container has three different session types. For the data published in June 2016 and October 2016, the last session is three_session_C</b<> while the data published after this were collected using three_session_C2. The key difference between these sessions is a change in the locally sparse noise stimulus. This is described more here.
Cell specimen ids#
During data processing, we matched identified ROIs across each of the sessions within experiment containers. Approximately one third of the neurons in the dataset were matched across all three sessions, one third were matched in two of the three session, and one third were only found in one session. Neurons have unique ids, called cell_specimen_ids, that are shared across the sessions they are found in.
How come we don’t always match ROIs across all three session for all neurons?
There are a few factors that could explain why we don’t always match ROIs across all sessions that include biological, experimental, and analytical reasons. Biologically, a neuron must be active within a session to be identifiable during segmentation. For various reasons, a neuron might not be active during some sessions while it is active during others. Experimentally, there are challenges to returning to the precise same field of view. Being at a slightly different depth, or having just a bit of tilt in the imaging plane, might result in some neurons that were in view during one session not being in view during another. Analytically, the method for identifying ROIs as well as for matching ROIs from multiple sessions can make mistakes.
We explore how to look at neurons across session in Cross session data.
