Single Cell Morphology#
Background#
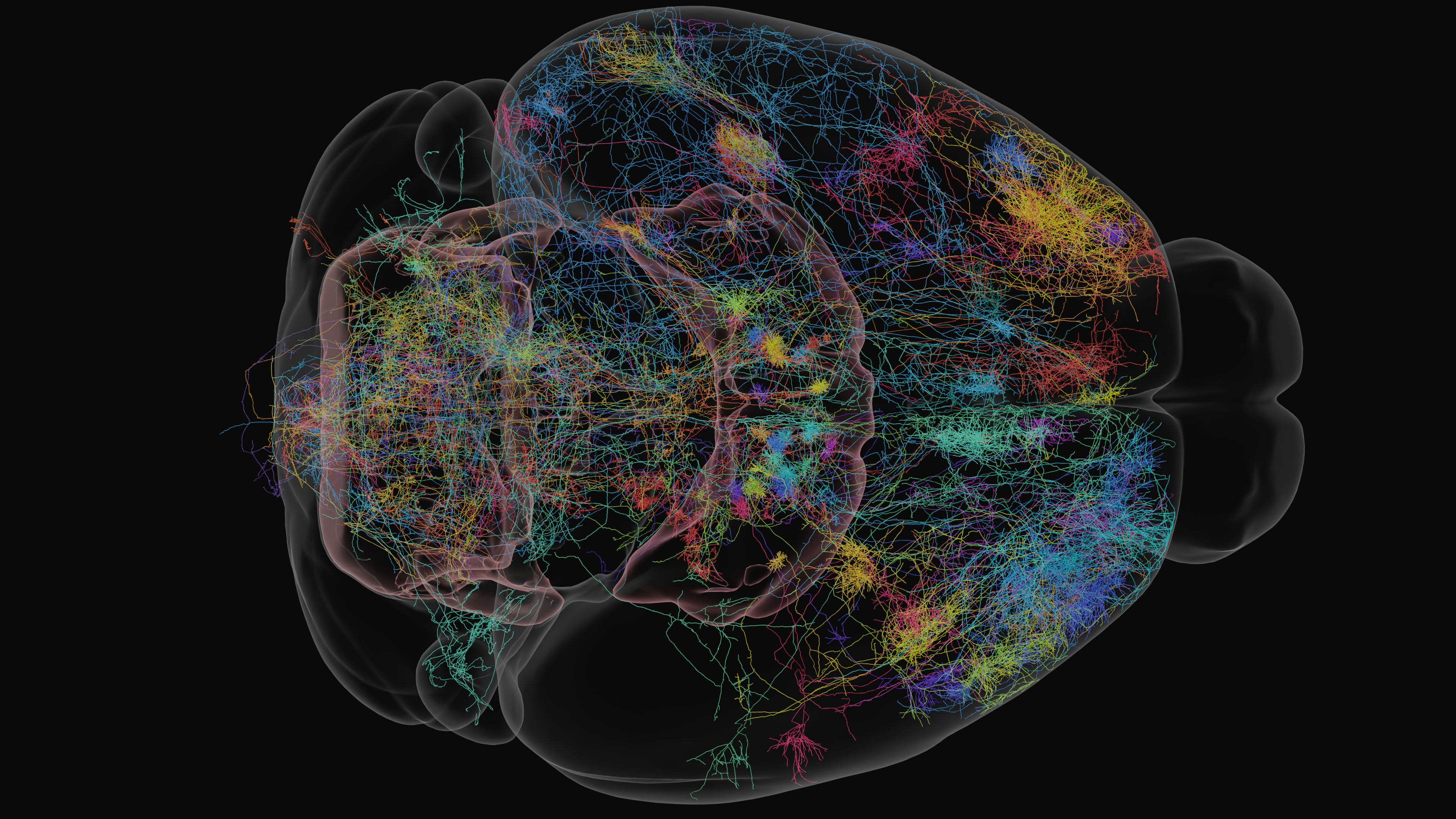
Delineating the connectivity within and between neural circuits is critical to understanding how information is routed through the brain. Neurons are the channels by which information flows and is transformed through the networks of the brain, but many neurons have a multiscale structure which makes brain-wide characterization difficult. Neurons can travel centimeter scale distances to target synaptic partners, but also possess important nanometer scale morphological features such as dendritic spines and axonal varicosities. The expansion-assisted selective plane illumination microscope (ExA-SPIM) system is designed to accommodate multiscale anatomy by dovetailing large field of view imaging with tissue clearing and expansion. This enables light-based imaging of entire mouse brains at nanoscale resolutions and subsequent fine-grained characterization of neuronal connectivity.
Whereas electron microscopy is used for dense reconstructions of small volumes of tissue, ExA-SPIM is employed to trace the long-range morphology of sparsely labelled neurons as they span the brain. For a given sample, a small number (ideally 30 – 100) of neurons are brightly labelled, and the whole brain is imaged. Neurons are then traced, either manually or in a semi-automated fashion, thereby reconstructing the neuron’s entire morphology. By registering data to a standardized template brain space (see Common Coordinate Framework section below), neuronal reconstructions from multiple brains can be analyzed in an integrated framework to understand morphological diversity and brain-wide connectivity. The data products contained within this dataset are single neurons described by sequences of points (also termed graphs, or skeletons) in this shared coordinate space, with metadata annotating the neuronal compartment (i.e. soma, axon, or dendrite) described by and brain region (e.g. primary visual cortex, mediodorsal nucleus of thalamus, dorsal striatum, etc.) occupied by that point.
What scientific questions can we Answer with this data?#
Some of the scientific questions that can be addressed with this dataset are as follows:
What do the inputs to a particular brain region look like? Where else do those inputs send collaterals?
What are the long-range outputs of a particular brain region? Are neuronal morphologies stereotyped, or are there distinct projection classes?
What is the morphological diversity of the cells with somas within a particular nucleus or brain area? Is any observed diversity discrete or continuous?
How do morphologies compare between different brain regions, e.g. higher order versus first order thalamus?
How do sub-compartments of a given neuronal type relate? Is dendritic morphology predictive of projection morphology?
