Accessing Neuropixels Visual Coding Data#
Tutorial overview#
This Jupyter notebook covers the various methods for accessing the Allen Institute Neuropixels Visual Coding dataset. We will go over how to request data, where it’s stored, and what the various files contain. If you’re having trouble downloading the data, or you just want to know more about what’s going on under the hood, this is a good place to start.
Currently, we do not have a web interface for browsing through the available cells and experiments, as with the two-photon imaging Visual Coding dataset. Instead, the data must be retrieved through the AllenSDK (Python 3.8+), or via requests sent to api.brain-map.org.
Functions related to data analysis will be covered in other tutorials. For a full list of available tutorials, see the SDK documentation.
This tutorial will cover the following topics:
Options for data access#
The EcephysProjectCache object of the AllenSDK is the easiest way to
interact with the data. This object abstracts away the details of on-disk file
storage, and delivers the data to you as ready-to-analyze Python objects. The
cache will automatically keep track of which files are stored locally, and will
download additional files on an as-needed basis. Usually you won’t need to worry
about how these files are structured, but this tutorial will cover those details
in case you want to analyze them without using the AllenSDK (e.g., in Matlab).
This tutorial begins with
an introduction to this approach.
If you have an Amazon Web Services (AWS) account, you can use an
EcephysProjectCache object to access the data via the Allen Brain Observatory
Simple Storage Service (S3) bucket. This is an AWS Public Dataset located at
arn:aws:s3:::allen-brain-observatory in region us-west-2. Launching a
Jupyter notebook instance on AWS will allow you to access the complete dataset
without having to download anything locally. This includes around 80 TB of raw
data files, which are not accessible via the AllenSDK. The only drawback is that
you’ll need to pay for the time that your instance is running—but this can still
be economical in many cases. A brief overview of this approach can be found
below.
A third option is to directly download the data via api.brain-map.org. This should be used only as a last resort if the other options are broken or are not available to you. Instructions for this can be found at the end of this tutorial.
Using the AllenSDK to retrieve data#
Most users will want to access data via the AllenSDK. This requires nothing more than a Python interpreter and some free disk space to store the data locally.
How much data is there? If you want to download the complete dataset (58 experiments), you’ll need 855 GB of space, split across the following files:
CSV files containing information about sessions, probes, channels and units (58.1 MB)
NWB files containing spike times, behavior data, and stimulus information for each session (146.5 GB total, min file size = 1.7 GB, max file size = 3.3 GB)
NWB files containing LFP data for each probe (707 GB total, min file size = 0.9 GB, max file size = 2.7 GB)
Before downloading the data, you must decide where the manifest.json file
lives. This file serves as the map that guides the EcephysProjectCache object
to the file locations.
When you initialize a local cache for the first time, it will create the manifest file at the path that you specify. This file lives in the same directory as the rest of the data, so make sure you put it somewhere that has enough space available.
When you need to access the data in subsequent analysis sessions, you should
point the EcephysProjectCache object to an existing manifest.json file;
otherwise, it will try to re-download the data in a new location.
To get started with this approach, first take care of the necessary imports:
import os
import shutil
import numpy as np
import pandas as pd
from pathlib import Path
from allensdk.brain_observatory.ecephys.ecephys_project_cache import EcephysProjectCache
/opt/envs/allensdk/lib/python3.10/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
Next, we’ll specify the location of the manifest file. If you’re creating a cache for the first time, this file won’t exist yet, but it must be placed in an existing data directory. Remember to choose a location that has plenty of free space available.
# Example cache directory path, it determines where downloaded data will be stored
output_dir = '/root/capsule/data/allen-brain-observatory/visual-coding-neuropixels/ecephys-cache/'
manifest_path = os.path.join(output_dir, 'manifest.json')
DOWNLOAD_COMPLETE_DATASET = True
Now we can create the cache object, specifying both the local storage directory (the manifest_path) and the remote storage location (the Allen Institute data warehouse).
cache = EcephysProjectCache.from_warehouse(manifest=manifest_path)
This will prepare the cache to download four files:
sessions.csv(7.8 kB)probes.csv(27.0 kB)channels.csv(6.6 MB)units.csv(51.4 MB)
Each one contains a table of information related to its file name. If you’re
using the AllenSDK, you won’t have to worry about how these files are formatted.
Instead, you’ll load the relevant data using specific accessor functions:
get_session_table(), get_probes(), get_channels(), and get_units().
These functions return a pandas
DataFrame
containing a row for each item and a column for each metric.
If you are analyzing data without using the AllenSDK, you can load the data using your CSV file reader of choice. However, please be aware the columns in the original file do not necessarily match what’s returned by the AllenSDK, which may combine information from multiple files to produce the final DataFrame.
Session metadata#
Let’s take a closer look at what’s in the sessions.csv file:
sessions = cache.get_session_table()
print('Total number of sessions: ' + str(len(sessions)))
Total number of sessions: 58
sessions.head()
| published_at | specimen_id | session_type | age_in_days | sex | full_genotype | unit_count | channel_count | probe_count | ecephys_structure_acronyms | |
|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||
| 715093703 | 2019-10-03T00:00:00Z | 699733581 | brain_observatory_1.1 | 118.0 | M | Sst-IRES-Cre/wt;Ai32(RCL-ChR2(H134R)_EYFP)/wt | 884 | 2219 | 6 | [CA1, VISrl, nan, PO, LP, LGd, CA3, DG, VISl, ... |
| 719161530 | 2019-10-03T00:00:00Z | 703279284 | brain_observatory_1.1 | 122.0 | M | Sst-IRES-Cre/wt;Ai32(RCL-ChR2(H134R)_EYFP)/wt | 755 | 2214 | 6 | [TH, Eth, APN, POL, LP, DG, CA1, VISpm, nan, N... |
| 721123822 | 2019-10-03T00:00:00Z | 707296982 | brain_observatory_1.1 | 125.0 | M | Pvalb-IRES-Cre/wt;Ai32(RCL-ChR2(H134R)_EYFP)/wt | 444 | 2229 | 6 | [MB, SCig, PPT, NOT, DG, CA1, VISam, nan, LP, ... |
| 732592105 | 2019-10-03T00:00:00Z | 717038288 | brain_observatory_1.1 | 100.0 | M | wt/wt | 824 | 1847 | 5 | [grey, VISpm, nan, VISp, VISl, VISal, VISrl] |
| 737581020 | 2019-10-03T00:00:00Z | 718643567 | brain_observatory_1.1 | 108.0 | M | wt/wt | 568 | 2218 | 6 | [grey, VISmma, nan, VISpm, VISp, VISl, VISrl] |
The sessions DataFrame provides a high-level overview of the Neuropixels
Visual Coding dataset. The index column is a unique ID, which serves as a key
for accessing the physiology data for each session. The other columns contain
information about:
the session type (i.e., which stimulus set was shown?)
the age, sex, and genotype of the mouse (in this dataset, there’s only one session per mouse)
the number of probes, channels, and units for each session
the brain structures recorded (CCFv3 acronyms)
As an illustrative example, if we want to find all of recordings from male Sst-Cre mice that viewed the Brain Observatory 1.1 stimulus and contain units from area LM, we can use the following query:
filtered_sessions = sessions[(sessions.sex == 'M') & \
(sessions.full_genotype.str.find('Sst') > -1) & \
(sessions.session_type == 'brain_observatory_1.1') & \
(['VISl' in acronyms for acronyms in
sessions.ecephys_structure_acronyms])]
filtered_sessions.head()
| published_at | specimen_id | session_type | age_in_days | sex | full_genotype | unit_count | channel_count | probe_count | ecephys_structure_acronyms | |
|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||
| 715093703 | 2019-10-03T00:00:00Z | 699733581 | brain_observatory_1.1 | 118.0 | M | Sst-IRES-Cre/wt;Ai32(RCL-ChR2(H134R)_EYFP)/wt | 884 | 2219 | 6 | [CA1, VISrl, nan, PO, LP, LGd, CA3, DG, VISl, ... |
| 719161530 | 2019-10-03T00:00:00Z | 703279284 | brain_observatory_1.1 | 122.0 | M | Sst-IRES-Cre/wt;Ai32(RCL-ChR2(H134R)_EYFP)/wt | 755 | 2214 | 6 | [TH, Eth, APN, POL, LP, DG, CA1, VISpm, nan, N... |
| 756029989 | 2019-10-03T00:00:00Z | 734865738 | brain_observatory_1.1 | 96.0 | M | Sst-IRES-Cre/wt;Ai32(RCL-ChR2(H134R)_EYFP)/wt | 684 | 2214 | 6 | [TH, DG, CA3, CA1, VISl, nan, PO, Eth, LP, VIS... |
The filtered_sessions table contains the three sessions that meet these
criteria.
The code above uses standard syntax for filtering pandas DataFrames. If this is unfamiliar to you, we strongly recommend reading through the pandas documentation. The AllenSDK makes heavy use of pandas objects, so we don’t have to come up with our own functions for working with tabular data.
The sessions DataFrame contains much of the basic information needed to
interpret the Visual Coding dataset. Below we demonstrate a few of these.
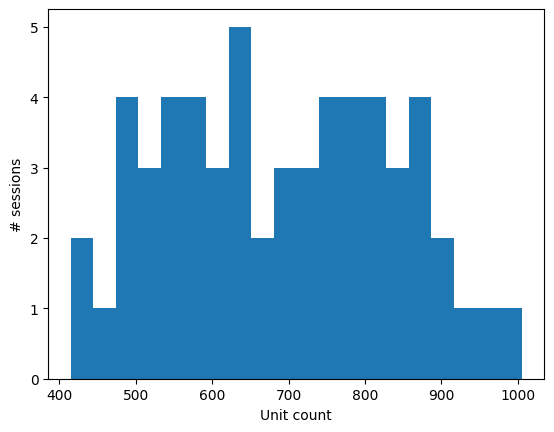
Number of units recorded in individual sessions:
print('Found {} sessions'.format(sessions.shape[0]))
print('Descriptive statistics for unit counts from individual sessions:')
print('Mean: {}'.format(sessions.unit_count.mean()))
print('Max: {}'.format(sessions.unit_count.max()))
print('Min: {}'.format(sessions.unit_count.min()))
Found 58 sessions
Descriptive statistics for unit counts from individual sessions:
Mean: 689.8275862068965
Max: 1005
Min: 415
import matplotlib.pyplot as plt
%matplotlib inline
plt.hist(sessions.unit_count, bins=20);
_ = plt.xlabel("Unit count")
_ = plt.ylabel("# sessions")
Genotypes of mice used in this dataset, and number of sessions available for each genotype:
sessions.full_genotype.value_counts()
wt/wt 30
Sst-IRES-Cre/wt;Ai32(RCL-ChR2(H134R)_EYFP)/wt 12
Pvalb-IRES-Cre/wt;Ai32(RCL-ChR2(H134R)_EYFP)/wt 8
Vip-IRES-Cre/wt;Ai32(RCL-ChR2(H134R)_EYFP)/wt 8
Name: full_genotype, dtype: int64
Types of sessions:
sessions.session_type.value_counts()
brain_observatory_1.1 32
functional_connectivity 26
Name: session_type, dtype: int64
CCFv3 acronyms of brain structures where the data were recorded:
all_areas = []
for index,row in sessions.iterrows():
for a in row.ecephys_structure_acronyms:
if a not in all_areas:
all_areas.append(a)
print(all_areas)
['CA1', 'VISrl', nan, 'PO', 'LP', 'LGd', 'CA3', 'DG', 'VISl', 'PoT', 'VISp', 'grey', 'VISpm', 'APN', 'MB', 'VISam', 'TH', 'Eth', 'POL', 'NOT', 'SUB', 'VL', 'CA2', 'VPM', 'VISal', 'SCig', 'PPT', 'VIS', 'ProS', 'LGv', 'HPF', 'VISmma', 'PP', 'PIL', 'MGv', 'VPL', 'IGL', 'SGN', 'IntG', 'LD', 'MGm', 'MGd', 'POST', 'MRN', 'VISli', 'OP', 'ZI', 'VISmmp', 'PF', 'LT', 'RPF', 'PRE', 'SCiw', 'CP', 'RT', 'SCop', 'SCsg', 'SCzo']
Number of sessions that have data from a given brain region (e.g. VISp):
count=0
for index,row in sessions.iterrows():
if 'VISp' in row.ecephys_structure_acronyms:
count+=1
print(count)
56
Probe metadata#
Let’s take a look at another DataFrame, extracted from the probes.csv file.
probes = cache.get_probes()
print('Total number of probes: ' + str(len(probes)))
probes.head()
Total number of probes: 332
| ecephys_session_id | lfp_sampling_rate | name | phase | sampling_rate | has_lfp_data | unit_count | channel_count | ecephys_structure_acronyms | |
|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||
| 729445648 | 719161530 | 1249.998642 | probeA | 3a | 29999.967418 | True | 87 | 374 | [APN, LP, MB, DG, CA1, VISam, nan] |
| 729445650 | 719161530 | 1249.996620 | probeB | 3a | 29999.918880 | True | 202 | 368 | [TH, Eth, APN, POL, LP, DG, CA1, VISpm, nan] |
| 729445652 | 719161530 | 1249.999897 | probeC | 3a | 29999.997521 | True | 207 | 373 | [APN, NOT, MB, DG, SUB, VISp, nan] |
| 729445654 | 719161530 | 1249.996707 | probeD | 3a | 29999.920963 | True | 93 | 358 | [grey, VL, CA3, CA2, CA1, VISl, nan] |
| 729445656 | 719161530 | 1249.999979 | probeE | 3a | 29999.999500 | True | 138 | 370 | [PO, VPM, TH, LP, LGd, CA3, DG, CA1, VISal, nan] |
The probes DataFrame contains information about the Neuropixels probes used
across all recordings. Each row represents one probe from one recording session,
even though the physical probes may have been used in multiple sessions. Some of
the important columns are:
ecephys_session_id: the index column of thesessionstablesampling_rate: the sampling rate (in Hz) for this probe’s spike band; note that each probe has a unique sampling rate around 30 kHz. The small variations in sampling rate across probes can add up to large offsets over time, so it’s critical to take these differences into account. However, all of the data you will interact with has been pre-aligned to a common clock, so this value is included only for reference purposes.lfp_sampling_rate: the sampling rate (in Hz) for this probe’s LFP band NWB files, after 2x downsampling from the original rate of 2.5 kHzname: the probe name is assigned based on the location of the probe on the recording rig. This is useful to keep in mind because probes with the same name are always targeted to the same cortical region and enter the brain from the same angle (probeA= AM,probeB= PM,probeC= V1,probeD= LM,probeE= AL,probeF= RL). However, the targeting is not always accurate, so the actual recorded region may be different.phase: the data may have been generated by one of two “phases” of Neuropixels probes. 3a = prototype version; PXI = publicly available version (“Neuropixels 1.0”). The two phases should be equivalent from the perspective of data analysis, but there may be differences in the noise characteristics between the two acquisition systems.channel_count: the number of channels with spikes or LFP data (maximum = 384)
Electrode channel metadata#
The channels.csv file contains information about each of these channels.
channels = cache.get_channels()
print('Total number of channels: ' + str(len(channels)))
channels.head()
Total number of channels: 123224
| ecephys_probe_id | local_index | probe_horizontal_position | probe_vertical_position | anterior_posterior_ccf_coordinate | dorsal_ventral_ccf_coordinate | left_right_ccf_coordinate | ecephys_structure_id | ecephys_structure_acronym | ecephys_session_id | lfp_sampling_rate | phase | sampling_rate | has_lfp_data | unit_count | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||
| 849705558 | 792645504 | 1 | 11 | 20 | 8165 | 3314 | 6862 | 215.0 | APN | 779839471 | 1250.001479 | 3a | 30000.035489 | True | 0 |
| 849705560 | 792645504 | 2 | 59 | 40 | 8162 | 3307 | 6866 | 215.0 | APN | 779839471 | 1250.001479 | 3a | 30000.035489 | True | 0 |
| 849705562 | 792645504 | 3 | 27 | 40 | 8160 | 3301 | 6871 | 215.0 | APN | 779839471 | 1250.001479 | 3a | 30000.035489 | True | 0 |
| 849705564 | 792645504 | 4 | 43 | 60 | 8157 | 3295 | 6875 | 215.0 | APN | 779839471 | 1250.001479 | 3a | 30000.035489 | True | 0 |
| 849705566 | 792645504 | 5 | 11 | 60 | 8155 | 3288 | 6879 | 215.0 | APN | 779839471 | 1250.001479 | 3a | 30000.035489 | True | 0 |
The most important columns in the channels DataFrame concern each channel’s
location in physical space. Each channel is associated with a location along the
probe shank (probe_horizontal_position and probe_vertical_position), and may
be linked to a coordinate in the Allen Common Coordinate framework (if CCF
registration is available for that probe).
The information about channel location will be merged into the units
DataFrame, which is loaded from units.csv:
Unit metadata#
units = cache.get_units()
print('Total number of units: ' + str(len(units)))
Total number of units: 40010
This DataFrame contains metadata about the available units across all sessions. By default, the AllenSDK applies some filters to this table and only returns units above a particular quality threshold.
The default filter values are as follows:
isi_violations< 0.5amplitude_cutoff< 0.1presence_ratio> 0.9
For more information about these quality metrics and how to interpret them, please refer to this tutorial.
If you want to see all of the available units, it’s straightforward to disable the quality metrics filters when retrieving this table:
units = cache.get_units(amplitude_cutoff_maximum = np.inf,
presence_ratio_minimum = -np.inf,
isi_violations_maximum = np.inf)
print('Total number of units: ' + str(len(units)))
Total number of units: 99180
units.head()
| waveform_PT_ratio | waveform_amplitude | amplitude_cutoff | cumulative_drift | d_prime | waveform_duration | ecephys_channel_id | firing_rate | waveform_halfwidth | isi_violations | ... | phase | sampling_rate | has_lfp_data | date_of_acquisition | published_at | specimen_id | session_type | age_in_days | sex | genotype | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||||
| 915956271 | 0.483286 | 53.203215 | 0.402024 | 271.58 | 3.484403 | 0.521943 | 850229415 | 10.562994 | 0.192295 | 0.122745 | ... | 3a | 29999.915391 | True | 2019-01-09T00:26:20Z | 2019-10-03T00:00:00Z | 717038288 | brain_observatory_1.1 | 100.0 | M | wt/wt |
| 915956287 | 0.463136 | 64.782510 | 0.500000 | 241.94 | 3.325401 | 0.618090 | 850229415 | 7.136036 | 0.274707 | 0.117272 | ... | 3a | 29999.915391 | True | 2019-01-09T00:26:20Z | 2019-10-03T00:00:00Z | 717038288 | brain_observatory_1.1 | 100.0 | M | wt/wt |
| 915956293 | 0.556859 | 112.967595 | 0.500000 | 308.46 | 2.496143 | 0.604355 | 850229415 | 5.066881 | 0.274707 | 0.279132 | ... | 3a | 29999.915391 | True | 2019-01-09T00:26:20Z | 2019-10-03T00:00:00Z | 717038288 | brain_observatory_1.1 | 100.0 | M | wt/wt |
| 915956282 | 0.611816 | 164.878740 | 0.072728 | 309.71 | 3.910873 | 0.535678 | 850229419 | 6.519432 | 0.164824 | 0.104910 | ... | 3a | 29999.915391 | True | 2019-01-09T00:26:20Z | 2019-10-03T00:00:00Z | 717038288 | brain_observatory_1.1 | 100.0 | M | wt/wt |
| 915956340 | 0.439372 | 247.254345 | 0.000881 | 160.24 | 5.519024 | 0.563149 | 850229419 | 9.660554 | 0.206030 | 0.006825 | ... | 3a | 29999.915391 | True | 2019-01-09T00:26:20Z | 2019-10-03T00:00:00Z | 717038288 | brain_observatory_1.1 | 100.0 | M | wt/wt |
5 rows × 45 columns
As you can see, the number of units has increased substantially, but some fraction of these units will be incomplete or highly contaminated. Understanding the meaning of these metrics is a critical part of analyzing the Neuropixels dataset, so we strongly recommend learning how to interpret them correctly.
Stimulus-specific unit metadata#
In addition to the quality metrics, there are a number of stimulus-specific metrics that are computed for each unit. These are not downloaded by default, but are accessed via a separate SDK function:
analysis_metrics1 = cache.get_unit_analysis_metrics_by_session_type('brain_observatory_1.1')
analysis_metrics2 = cache.get_unit_analysis_metrics_by_session_type('functional_connectivity')
print(str(len(analysis_metrics1)) + ' units in table 1')
print(str(len(analysis_metrics2)) + ' units in table 2')
21842 units in table 1
18168 units in table 2
This will download two additional files,
brain_observatory_1.1_analysis_metrics.csv and
functional_connectivity_analysis_metrics.csv, and load them as pandas
DataFrames. Note that the total length of these DataFrames is around 40k units,
because the default quality metric thresholds have been applied.
To load all of the available units, and create one giant table of metrics, you can use the following code:
analysis_metrics1 = cache.get_unit_analysis_metrics_by_session_type(
'brain_observatory_1.1',
amplitude_cutoff_maximum = np.inf,
presence_ratio_minimum = -np.inf,
isi_violations_maximum = np.inf
)
analysis_metrics2 = cache.get_unit_analysis_metrics_by_session_type(
'functional_connectivity',
amplitude_cutoff_maximum = np.inf,
presence_ratio_minimum = -np.inf,
isi_violations_maximum = np.inf
)
all_metrics = pd.concat([analysis_metrics1, analysis_metrics2], sort=False)
print(str(len(all_metrics)) + ' units overall')
99180 units overall
The length of this DataFrame should match that of the units DataFrame we
retrieved earlier. A few things to note about this DataFrame:
The unit analysis metrics DataFrame also includes all quality metrics, so it’s a superset of the
unitsDataFrameSince some of the stimuli in the
brain_observatory_1.1session are not present in thefunctional_connectivitysession, many of the data points in the unit analysis metrics DataFrame will be filled withnanvalues
Data for individual sessions#
Assuming you’ve found a session you’re interested in analyzing in more detail,
it’s now time to download the data. This is as simple as calling
cache.get_session_data(), with the session_id as input. This method will
check the cache for an existing NWB file and, if it’s not present, will
automatically download it for you.
Each NWB file can be upwards of 2 GB, so please be patient while it’s downloading!
As an example, let’s look at one of the sessions we selected earlier, disabling the default unit quality metrics filters:
session = cache.get_session_data(filtered_sessions.index.values[0],
isi_violations_maximum = np.inf,
amplitude_cutoff_maximum = np.inf,
presence_ratio_minimum = -np.inf
)
print([attr_or_method for attr_or_method in dir(session) if attr_or_method[0] != '_'])
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'hdmf-common' version 1.1.3 because version 1.8.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.2.2 because version 2.7.0 is already loaded.
return func(args[0], **pargs)
['DETAILED_STIMULUS_PARAMETERS', 'LazyProperty', 'age_in_days', 'api', 'channel_structure_intervals', 'channels', 'conditionwise_spike_statistics', 'ecephys_session_id', 'from_nwb_path', 'full_genotype', 'get_current_source_density', 'get_inter_presentation_intervals_for_stimulus', 'get_invalid_times', 'get_lfp', 'get_parameter_values_for_stimulus', 'get_pupil_data', 'get_screen_gaze_data', 'get_stimulus_epochs', 'get_stimulus_parameter_values', 'get_stimulus_table', 'inter_presentation_intervals', 'invalid_times', 'mean_waveforms', 'metadata', 'num_channels', 'num_probes', 'num_stimulus_presentations', 'num_units', 'optogenetic_stimulation_epochs', 'presentationwise_spike_counts', 'presentationwise_spike_times', 'probes', 'rig_equipment_name', 'rig_geometry_data', 'running_speed', 'session_start_time', 'session_type', 'sex', 'specimen_name', 'spike_amplitudes', 'spike_times', 'stimulus_conditions', 'stimulus_names', 'stimulus_presentations', 'structure_acronyms', 'structurewise_unit_counts', 'units']
As you can see, the session object has a lot of attributes and methods that
can be used to access the underlying data in the NWB file. Most of these will be
touched on in other tutorials, but for now we will look at the only one that is
capable of triggering additional data downloads, get_lfp().
In general, each NWB file is meant to be a self-contained repository of data for one recording session. However, for the Neuropixels data, we’ve broken with convention a bit in order to store LFP data in separate files. If we hadn’t done this, analyzing one session would require an initial 15 GB file download. Now, the session is broken up in to ~2 GB chunks..
Once you have created a session object, downloading the LFP data is simple
(but may be slow):
probe_id = session.probes.index.values[0]
lfp = session.get_lfp(probe_id)
Tips for analyzing LFP data can be found in this tutorial.
Downloading the complete dataset#
Analyzing one session at a time is nice, but in many case you’ll want to be able
to query across the whole dataset. To fill your cache with all available data,
you can use a for loop like the one below. Note that we’ve added some checks
to ensure that the complete file is present, in case the download has been
interrupted due to an unreliable connection.
Before running this code, please make sure that you have enough space available in your cache directory. You’ll need around 855 GB for the whole dataset, and 147 GB if you’re not downloading the LFP data files.
if DOWNLOAD_COMPLETE_DATASET:
for session_id, row in sessions.iterrows():
truncated_file = True
directory = os.path.join(output_dir + '/session_' + str(session_id))
while truncated_file:
session = cache.get_session_data(session_id)
try:
print(session.specimen_name)
truncated_file = False
except OSError:
shutil.rmtree(directory)
print(" Truncated spikes file, re-downloading")
for probe_id, probe in session.probes.iterrows():
print(' ' + probe.description)
truncated_lfp = True
while truncated_lfp:
try:
lfp = session.get_lfp(probe_id)
truncated_lfp = False
except OSError:
fname = directory + '/probe_' + str(probe_id) + '_lfp.nwb'
os.remove(fname)
print(" Truncated LFP file, re-downloading")
except ValueError:
print(" LFP file not found.")
truncated_lfp = False
Accessing data on AWS#
If you want to analyze the data without downloading anything to your local machine, you can use the AllenSDK on AWS.
Follow these instructions to launch a Jupyter notebook. Then, simply point to the existing manifest file in the Allen Institute’s S3 bucket, and all of the data will be immediately available:
cache = EcephysProjectCache(manifest=manifest_path)
Once your cache is initialized, you can create the sessions table, load
individual session objects, and access LFP data using the same commands
described above.
Additional tutorials specific to using AWS are coming soon.
Direct download via api.brain-map.org#
Some people have reported issues downloading the files via the AllenSDK (the connection is extremely slow, or gets interrupted frequently). If this applies to you, you can try downloading the files via HTTP requests sent to api.brain-map.org. This approach is not recommended, because you will have to manually keep track of the file locations. But if you’re doing analysis that doesn’t depend on the AllenSDK (e.g., in Matlab), this may not matter to you.
You can follow the steps below to retrieve the URLs for all of the NWB files in this dataset.
from allensdk.brain_observatory.ecephys.ecephys_project_api.utilities import build_and_execute
from allensdk.brain_observatory.ecephys.ecephys_project_api.rma_engine import RmaEngine
from allensdk.brain_observatory.ecephys.ecephys_project_cache import EcephysProjectCache
rma_engine = RmaEngine(scheme="http", host="api.brain-map.org")
cache = EcephysProjectCache.from_warehouse(manifest=manifest_path)
sessions = cache.get_session_table()
def retrieve_link(session_id):
well_known_files = build_and_execute(
(
"criteria=model::WellKnownFile"
",rma::criteria,well_known_file_type[name$eq'EcephysNwb']"
"[attachable_type$eq'EcephysSession']"
r"[attachable_id$eq{{session_id}}]"
),
engine=rma_engine.get_rma_tabular,
session_id=session_id
)
return 'http://api.brain-map.org/' + well_known_files['download_link'].iloc[0]
download_links = [retrieve_link(session_id) for session_id in sessions.index.values]
_ = [print(link) for link in download_links]
http://api.brain-map.org//api/v2/well_known_file_download/1026124469
http://api.brain-map.org//api/v2/well_known_file_download/1026124034
http://api.brain-map.org//api/v2/well_known_file_download/1026123696
http://api.brain-map.org//api/v2/well_known_file_download/1026123599
http://api.brain-map.org//api/v2/well_known_file_download/1026123989
http://api.brain-map.org//api/v2/well_known_file_download/1026123897
http://api.brain-map.org//api/v2/well_known_file_download/1026123964
http://api.brain-map.org//api/v2/well_known_file_download/1026124068
http://api.brain-map.org//api/v2/well_known_file_download/1026124429
http://api.brain-map.org//api/v2/well_known_file_download/1026124262
http://api.brain-map.org//api/v2/well_known_file_download/1026124724
http://api.brain-map.org//api/v2/well_known_file_download/1026124242
http://api.brain-map.org//api/v2/well_known_file_download/1026124863
http://api.brain-map.org//api/v2/well_known_file_download/1026123537
http://api.brain-map.org//api/v2/well_known_file_download/1026124326
http://api.brain-map.org//api/v2/well_known_file_download/1026124684
http://api.brain-map.org//api/v2/well_known_file_download/1026124216
http://api.brain-map.org//api/v2/well_known_file_download/1026124603
http://api.brain-map.org//api/v2/well_known_file_download/1026123685
http://api.brain-map.org//api/v2/well_known_file_download/1026123877
http://api.brain-map.org//api/v2/well_known_file_download/1026123377
http://api.brain-map.org//api/v2/well_known_file_download/1026123651
http://api.brain-map.org//api/v2/well_known_file_download/1026124702
http://api.brain-map.org//api/v2/well_known_file_download/1026124793
http://api.brain-map.org//api/v2/well_known_file_download/1026124884
http://api.brain-map.org//api/v2/well_known_file_download/1026124569
http://api.brain-map.org//api/v2/well_known_file_download/1026124500
http://api.brain-map.org//api/v2/well_known_file_download/1026124293
http://api.brain-map.org//api/v2/well_known_file_download/1026124194
http://api.brain-map.org//api/v2/well_known_file_download/1026124517
http://api.brain-map.org//api/v2/well_known_file_download/1026124918
http://api.brain-map.org//api/v2/well_known_file_download/1026124152
http://api.brain-map.org//api/v2/well_known_file_download/1026124109
http://api.brain-map.org//api/v2/well_known_file_download/1026124348
http://api.brain-map.org//api/v2/well_known_file_download/1026124625
http://api.brain-map.org//api/v2/well_known_file_download/1026124179
http://api.brain-map.org//api/v2/well_known_file_download/1026124007
http://api.brain-map.org//api/v2/well_known_file_download/1026124309
http://api.brain-map.org//api/v2/well_known_file_download/1026123803
http://api.brain-map.org//api/v2/well_known_file_download/1026124405
http://api.brain-map.org//api/v2/well_known_file_download/1026123847
http://api.brain-map.org//api/v2/well_known_file_download/1026124645
http://api.brain-map.org//api/v2/well_known_file_download/1026124545
http://api.brain-map.org//api/v2/well_known_file_download/1026124759
http://api.brain-map.org//api/v2/well_known_file_download/1026124804
http://api.brain-map.org//api/v2/well_known_file_download/1026124743
http://api.brain-map.org//api/v2/well_known_file_download/1026125021
http://api.brain-map.org//api/v2/well_known_file_download/1026124840
http://api.brain-map.org//api/v2/well_known_file_download/1026124085
http://api.brain-map.org//api/v2/well_known_file_download/1026124368
http://api.brain-map.org//api/v2/well_known_file_download/1026123824
http://api.brain-map.org//api/v2/well_known_file_download/1026123625
http://api.brain-map.org//api/v2/well_known_file_download/1026123787
http://api.brain-map.org//api/v2/well_known_file_download/1026123519
http://api.brain-map.org//api/v2/well_known_file_download/1026123725
http://api.brain-map.org//api/v2/well_known_file_download/1026122596
http://api.brain-map.org//api/v2/well_known_file_download/1026124422
http://api.brain-map.org//api/v2/well_known_file_download/1026123943
download_links is a list of 58 links that can be used to download the NWB
files for all available sessions. Clicking on the links above should start the
download automatically.
Please keep in mind that you’ll have to move these files to the appropriate
sub-directory once the download is complete. The EcephysProjectCache object
expects the following directory structure:
cache_dir/
+-- manifest.json
+-- session_<id>/
¦ +-- session_<id>.nwb
+-- session_<id>/
¦ +-- session_<id>.nwb
+-- session_<id>/
¦ +-- session_<id>.nwb
If you aren’t interested in using the EcephysProjectCache object to keep track
of what you’ve downloaded, you can create a session object just by passing a
path to an NWB file:
from allensdk.brain_observatory.ecephys.ecephys_session import EcephysSession
nwb_path = '/mnt/nvme0/ecephys_cache_dir_10_31/session_721123822/session_721123822.nwb'
session = EcephysSession.from_nwb_path(nwb_path, api_kwargs={
"amplitude_cutoff_maximum": np.inf,
"presence_ratio_minimum": -np.inf,
"isi_violations_maximum": np.inf
})
This will load the data for one session, without applying the default unit
quality metric filters. Everything will be available except the LFP data,
because the get_lfp() method can only find the associated LFP files if you’re
using the EcephysProjectCache object.
To obtain similar links for the LFP files, you can use the following code:
def retrieve_lfp_link(probe_id):
well_known_files = build_and_execute(
(
"criteria=model::WellKnownFile"
",rma::criteria,well_known_file_type[name$eq'EcephysLfpNwb']"
"[attachable_type$eq'EcephysProbe']"
r"[attachable_id$eq{{probe_id}}]"
),
engine=rma_engine.get_rma_tabular,
probe_id=probe_id
)
if well_known_files.shape[0] != 1:
return 'file for probe ' + str(probe_id) + ' not found'
return 'http://api.brain-map.org/' + well_known_files.loc[0, "download_link"]
probes = cache.get_probes()
download_links = [retrieve_lfp_link(probe_id) for probe_id in probes.index.values]
_ = [print(link) for link in download_links]
http://api.brain-map.org//api/v2/well_known_file_download/1026124040
http://api.brain-map.org//api/v2/well_known_file_download/1026124036
http://api.brain-map.org//api/v2/well_known_file_download/1026124038
http://api.brain-map.org//api/v2/well_known_file_download/1026124042
http://api.brain-map.org//api/v2/well_known_file_download/1026124044
http://api.brain-map.org//api/v2/well_known_file_download/1026124046
http://api.brain-map.org//api/v2/well_known_file_download/1026123601
http://api.brain-map.org//api/v2/well_known_file_download/1026123603
http://api.brain-map.org//api/v2/well_known_file_download/1026123605
http://api.brain-map.org//api/v2/well_known_file_download/1026123607
http://api.brain-map.org//api/v2/well_known_file_download/1026123609
http://api.brain-map.org//api/v2/well_known_file_download/1026123539
http://api.brain-map.org//api/v2/well_known_file_download/1026123543
http://api.brain-map.org//api/v2/well_known_file_download/1026123550
http://api.brain-map.org//api/v2/well_known_file_download/1026123546
http://api.brain-map.org//api/v2/well_known_file_download/1026123541
http://api.brain-map.org//api/v2/well_known_file_download/1026123548
http://api.brain-map.org//api/v2/well_known_file_download/1026124433
http://api.brain-map.org//api/v2/well_known_file_download/1026124436
http://api.brain-map.org//api/v2/well_known_file_download/1026124439
http://api.brain-map.org//api/v2/well_known_file_download/1026124442
http://api.brain-map.org//api/v2/well_known_file_download/1026124446
http://api.brain-map.org//api/v2/well_known_file_download/1026124448
http://api.brain-map.org//api/v2/well_known_file_download/1026124728
http://api.brain-map.org//api/v2/well_known_file_download/1026124726
http://api.brain-map.org//api/v2/well_known_file_download/1026124730
http://api.brain-map.org//api/v2/well_known_file_download/1026124732
http://api.brain-map.org//api/v2/well_known_file_download/1026124737
http://api.brain-map.org//api/v2/well_known_file_download/1026124735
http://api.brain-map.org//api/v2/well_known_file_download/1026123901
http://api.brain-map.org//api/v2/well_known_file_download/1026123909
http://api.brain-map.org//api/v2/well_known_file_download/1026123907
http://api.brain-map.org//api/v2/well_known_file_download/1026123905
http://api.brain-map.org//api/v2/well_known_file_download/1026123903
http://api.brain-map.org//api/v2/well_known_file_download/1026123899
http://api.brain-map.org//api/v2/well_known_file_download/1026124867
http://api.brain-map.org//api/v2/well_known_file_download/1026124871
http://api.brain-map.org//api/v2/well_known_file_download/1026124873
http://api.brain-map.org//api/v2/well_known_file_download/1026124875
http://api.brain-map.org//api/v2/well_known_file_download/1026124869
http://api.brain-map.org//api/v2/well_known_file_download/1026124865
http://api.brain-map.org//api/v2/well_known_file_download/1026123993
http://api.brain-map.org//api/v2/well_known_file_download/1026123997
http://api.brain-map.org//api/v2/well_known_file_download/1026124001
http://api.brain-map.org//api/v2/well_known_file_download/1026123999
http://api.brain-map.org//api/v2/well_known_file_download/1026123995
http://api.brain-map.org//api/v2/well_known_file_download/1026123991
http://api.brain-map.org//api/v2/well_known_file_download/1026123702
http://api.brain-map.org//api/v2/well_known_file_download/1026123704
http://api.brain-map.org//api/v2/well_known_file_download/1026123700
http://api.brain-map.org//api/v2/well_known_file_download/1026123706
http://api.brain-map.org//api/v2/well_known_file_download/1026123710
http://api.brain-map.org//api/v2/well_known_file_download/1026123708
http://api.brain-map.org//api/v2/well_known_file_download/1026124330
http://api.brain-map.org//api/v2/well_known_file_download/1026124334
http://api.brain-map.org//api/v2/well_known_file_download/1026124337
http://api.brain-map.org//api/v2/well_known_file_download/1026124332
http://api.brain-map.org//api/v2/well_known_file_download/1026124328
http://api.brain-map.org//api/v2/well_known_file_download/1026124229
http://api.brain-map.org//api/v2/well_known_file_download/1026124227
http://api.brain-map.org//api/v2/well_known_file_download/1026124225
http://api.brain-map.org//api/v2/well_known_file_download/1026124221
http://api.brain-map.org//api/v2/well_known_file_download/1026124223
http://api.brain-map.org//api/v2/well_known_file_download/1026124219
http://api.brain-map.org//api/v2/well_known_file_download/1026124697
http://api.brain-map.org//api/v2/well_known_file_download/1026124695
http://api.brain-map.org//api/v2/well_known_file_download/1026124688
http://api.brain-map.org//api/v2/well_known_file_download/1026124686
http://api.brain-map.org//api/v2/well_known_file_download/1026124690
http://api.brain-map.org//api/v2/well_known_file_download/1026124693
http://api.brain-map.org//api/v2/well_known_file_download/1026124268
http://api.brain-map.org//api/v2/well_known_file_download/1026124266
http://api.brain-map.org//api/v2/well_known_file_download/1026124264
http://api.brain-map.org//api/v2/well_known_file_download/1026124270
http://api.brain-map.org//api/v2/well_known_file_download/1026124272
http://api.brain-map.org//api/v2/well_known_file_download/1026124274
http://api.brain-map.org//api/v2/well_known_file_download/1026124888
http://api.brain-map.org//api/v2/well_known_file_download/1026124890
http://api.brain-map.org//api/v2/well_known_file_download/1026124886
http://api.brain-map.org//api/v2/well_known_file_download/1026124892
http://api.brain-map.org//api/v2/well_known_file_download/1026124894
http://api.brain-map.org//api/v2/well_known_file_download/1026124803
http://api.brain-map.org//api/v2/well_known_file_download/1026124799
http://api.brain-map.org//api/v2/well_known_file_download/1026124795
http://api.brain-map.org//api/v2/well_known_file_download/1026124801
http://api.brain-map.org//api/v2/well_known_file_download/1026124797
http://api.brain-map.org//api/v2/well_known_file_download/1026124806
http://api.brain-map.org//api/v2/well_known_file_download/1026124571
http://api.brain-map.org//api/v2/well_known_file_download/1026124575
http://api.brain-map.org//api/v2/well_known_file_download/1026124579
http://api.brain-map.org//api/v2/well_known_file_download/1026124577
http://api.brain-map.org//api/v2/well_known_file_download/1026124582
http://api.brain-map.org//api/v2/well_known_file_download/1026124573
http://api.brain-map.org//api/v2/well_known_file_download/1026123975
http://api.brain-map.org//api/v2/well_known_file_download/1026123978
http://api.brain-map.org//api/v2/well_known_file_download/1026123973
http://api.brain-map.org//api/v2/well_known_file_download/1026123966
http://api.brain-map.org//api/v2/well_known_file_download/1026123971
http://api.brain-map.org//api/v2/well_known_file_download/1026123969
http://api.brain-map.org//api/v2/well_known_file_download/1026124614
http://api.brain-map.org//api/v2/well_known_file_download/1026124612
http://api.brain-map.org//api/v2/well_known_file_download/1026124616
http://api.brain-map.org//api/v2/well_known_file_download/1026124610
http://api.brain-map.org//api/v2/well_known_file_download/1026124607
http://api.brain-map.org//api/v2/well_known_file_download/1026124605
http://api.brain-map.org//api/v2/well_known_file_download/1026123385
http://api.brain-map.org//api/v2/well_known_file_download/1026123383
http://api.brain-map.org//api/v2/well_known_file_download/1026123379
http://api.brain-map.org//api/v2/well_known_file_download/1026123381
http://api.brain-map.org//api/v2/well_known_file_download/1026124248
http://api.brain-map.org//api/v2/well_known_file_download/1026124246
http://api.brain-map.org//api/v2/well_known_file_download/1026124252
http://api.brain-map.org//api/v2/well_known_file_download/1026124250
http://api.brain-map.org//api/v2/well_known_file_download/1026124254
http://api.brain-map.org//api/v2/well_known_file_download/1026124244
http://api.brain-map.org//api/v2/well_known_file_download/1026124709
http://api.brain-map.org//api/v2/well_known_file_download/1026124707
http://api.brain-map.org//api/v2/well_known_file_download/1026124704
http://api.brain-map.org//api/v2/well_known_file_download/1026124711
http://api.brain-map.org//api/v2/well_known_file_download/1026124713
http://api.brain-map.org//api/v2/well_known_file_download/1026124715
http://api.brain-map.org//api/v2/well_known_file_download/1026123691
http://api.brain-map.org//api/v2/well_known_file_download/1026123695
http://api.brain-map.org//api/v2/well_known_file_download/1026123693
http://api.brain-map.org//api/v2/well_known_file_download/1026123689
http://api.brain-map.org//api/v2/well_known_file_download/1026123687
http://api.brain-map.org//api/v2/well_known_file_download/1026124073
http://api.brain-map.org//api/v2/well_known_file_download/1026124075
http://api.brain-map.org//api/v2/well_known_file_download/1026124071
http://api.brain-map.org//api/v2/well_known_file_download/1026124082
http://api.brain-map.org//api/v2/well_known_file_download/1026124079
http://api.brain-map.org//api/v2/well_known_file_download/1026124077
http://api.brain-map.org//api/v2/well_known_file_download/1026123879
http://api.brain-map.org//api/v2/well_known_file_download/1026123881
http://api.brain-map.org//api/v2/well_known_file_download/1026123883
http://api.brain-map.org//api/v2/well_known_file_download/1026123885
http://api.brain-map.org//api/v2/well_known_file_download/1026124205
http://api.brain-map.org//api/v2/well_known_file_download/1026124208
http://api.brain-map.org//api/v2/well_known_file_download/1026124199
http://api.brain-map.org//api/v2/well_known_file_download/1026124197
http://api.brain-map.org//api/v2/well_known_file_download/1026124201
http://api.brain-map.org//api/v2/well_known_file_download/1026124203
http://api.brain-map.org//api/v2/well_known_file_download/1026124507
http://api.brain-map.org//api/v2/well_known_file_download/1026124504
http://api.brain-map.org//api/v2/well_known_file_download/1026124502
http://api.brain-map.org//api/v2/well_known_file_download/1026124509
file for probe 773463023 not found
http://api.brain-map.org//api/v2/well_known_file_download/1026124511
http://api.brain-map.org//api/v2/well_known_file_download/1026124519
http://api.brain-map.org//api/v2/well_known_file_download/1026124523
http://api.brain-map.org//api/v2/well_known_file_download/1026124527
http://api.brain-map.org//api/v2/well_known_file_download/1026124525
http://api.brain-map.org//api/v2/well_known_file_download/1026124521
http://api.brain-map.org//api/v2/well_known_file_download/1026124529
http://api.brain-map.org//api/v2/well_known_file_download/1026124299
http://api.brain-map.org//api/v2/well_known_file_download/1026124297
http://api.brain-map.org//api/v2/well_known_file_download/1026124295
http://api.brain-map.org//api/v2/well_known_file_download/1026124301
http://api.brain-map.org//api/v2/well_known_file_download/1026124303
http://api.brain-map.org//api/v2/well_known_file_download/1026124305
http://api.brain-map.org//api/v2/well_known_file_download/1026124922
http://api.brain-map.org//api/v2/well_known_file_download/1026124924
http://api.brain-map.org//api/v2/well_known_file_download/1026124926
http://api.brain-map.org//api/v2/well_known_file_download/1026124928
http://api.brain-map.org//api/v2/well_known_file_download/1026124930
http://api.brain-map.org//api/v2/well_known_file_download/1026124920
http://api.brain-map.org//api/v2/well_known_file_download/1026124156
http://api.brain-map.org//api/v2/well_known_file_download/1026124164
http://api.brain-map.org//api/v2/well_known_file_download/1026124160
http://api.brain-map.org//api/v2/well_known_file_download/1026124162
http://api.brain-map.org//api/v2/well_known_file_download/1026124158
http://api.brain-map.org//api/v2/well_known_file_download/1026124154
http://api.brain-map.org//api/v2/well_known_file_download/1026124415
http://api.brain-map.org//api/v2/well_known_file_download/1026124413
http://api.brain-map.org//api/v2/well_known_file_download/1026124417
http://api.brain-map.org//api/v2/well_known_file_download/1026124409
http://api.brain-map.org//api/v2/well_known_file_download/1026124411
http://api.brain-map.org//api/v2/well_known_file_download/1026124407
http://api.brain-map.org//api/v2/well_known_file_download/1026124311
http://api.brain-map.org//api/v2/well_known_file_download/1026124313
http://api.brain-map.org//api/v2/well_known_file_download/1026124319
http://api.brain-map.org//api/v2/well_known_file_download/1026124317
http://api.brain-map.org//api/v2/well_known_file_download/1026124321
http://api.brain-map.org//api/v2/well_known_file_download/1026124315
http://api.brain-map.org//api/v2/well_known_file_download/1026124350
http://api.brain-map.org//api/v2/well_known_file_download/1026124354
http://api.brain-map.org//api/v2/well_known_file_download/1026124356
http://api.brain-map.org//api/v2/well_known_file_download/1026124358
http://api.brain-map.org//api/v2/well_known_file_download/1026124360
http://api.brain-map.org//api/v2/well_known_file_download/1026124352
http://api.brain-map.org//api/v2/well_known_file_download/1026124627
http://api.brain-map.org//api/v2/well_known_file_download/1026124631
http://api.brain-map.org//api/v2/well_known_file_download/1026124629
http://api.brain-map.org//api/v2/well_known_file_download/1026124634
http://api.brain-map.org//api/v2/well_known_file_download/1026124638
http://api.brain-map.org//api/v2/well_known_file_download/1026124636
http://api.brain-map.org//api/v2/well_known_file_download/1026123805
http://api.brain-map.org//api/v2/well_known_file_download/1026123809
http://api.brain-map.org//api/v2/well_known_file_download/1026123807
http://api.brain-map.org//api/v2/well_known_file_download/1026123811
http://api.brain-map.org//api/v2/well_known_file_download/1026123813
http://api.brain-map.org//api/v2/well_known_file_download/1026123815
http://api.brain-map.org//api/v2/well_known_file_download/1026124183
http://api.brain-map.org//api/v2/well_known_file_download/1026124187
http://api.brain-map.org//api/v2/well_known_file_download/1026124185
http://api.brain-map.org//api/v2/well_known_file_download/1026124181
http://api.brain-map.org//api/v2/well_known_file_download/1026124189
http://api.brain-map.org//api/v2/well_known_file_download/1026124191
http://api.brain-map.org//api/v2/well_known_file_download/1026124013
http://api.brain-map.org//api/v2/well_known_file_download/1026124011
http://api.brain-map.org//api/v2/well_known_file_download/1026124009
http://api.brain-map.org//api/v2/well_known_file_download/1026124015
http://api.brain-map.org//api/v2/well_known_file_download/1026124017
http://api.brain-map.org//api/v2/well_known_file_download/1026124019
http://api.brain-map.org//api/v2/well_known_file_download/1026124111
http://api.brain-map.org//api/v2/well_known_file_download/1026124116
http://api.brain-map.org//api/v2/well_known_file_download/1026124114
http://api.brain-map.org//api/v2/well_known_file_download/1026124118
http://api.brain-map.org//api/v2/well_known_file_download/1026124120
http://api.brain-map.org//api/v2/well_known_file_download/1026124123
http://api.brain-map.org//api/v2/well_known_file_download/1026124754
http://api.brain-map.org//api/v2/well_known_file_download/1026124748
http://api.brain-map.org//api/v2/well_known_file_download/1026124746
http://api.brain-map.org//api/v2/well_known_file_download/1026124752
http://api.brain-map.org//api/v2/well_known_file_download/1026124756
http://api.brain-map.org//api/v2/well_known_file_download/1026124750
http://api.brain-map.org//api/v2/well_known_file_download/1026123849
http://api.brain-map.org//api/v2/well_known_file_download/1026123857
http://api.brain-map.org//api/v2/well_known_file_download/1026123853
http://api.brain-map.org//api/v2/well_known_file_download/1026123851
http://api.brain-map.org//api/v2/well_known_file_download/1026123859
http://api.brain-map.org//api/v2/well_known_file_download/1026123855
http://api.brain-map.org//api/v2/well_known_file_download/1026124653
http://api.brain-map.org//api/v2/well_known_file_download/1026124651
http://api.brain-map.org//api/v2/well_known_file_download/1026124657
http://api.brain-map.org//api/v2/well_known_file_download/1026124647
http://api.brain-map.org//api/v2/well_known_file_download/1026124649
http://api.brain-map.org//api/v2/well_known_file_download/1026124655
http://api.brain-map.org//api/v2/well_known_file_download/1026124553
http://api.brain-map.org//api/v2/well_known_file_download/1026124551
http://api.brain-map.org//api/v2/well_known_file_download/1026124549
http://api.brain-map.org//api/v2/well_known_file_download/1026124547
http://api.brain-map.org//api/v2/well_known_file_download/1026124555
http://api.brain-map.org//api/v2/well_known_file_download/1026124557
http://api.brain-map.org//api/v2/well_known_file_download/1026125023
http://api.brain-map.org//api/v2/well_known_file_download/1026125027
http://api.brain-map.org//api/v2/well_known_file_download/1026125025
http://api.brain-map.org//api/v2/well_known_file_download/1026125029
http://api.brain-map.org//api/v2/well_known_file_download/1026125031
http://api.brain-map.org//api/v2/well_known_file_download/1026125034
http://api.brain-map.org//api/v2/well_known_file_download/1026124814
http://api.brain-map.org//api/v2/well_known_file_download/1026124808
http://api.brain-map.org//api/v2/well_known_file_download/1026124811
http://api.brain-map.org//api/v2/well_known_file_download/1026124816
http://api.brain-map.org//api/v2/well_known_file_download/1026124820
http://api.brain-map.org//api/v2/well_known_file_download/1026124818
http://api.brain-map.org//api/v2/well_known_file_download/1026123659
http://api.brain-map.org//api/v2/well_known_file_download/1026123657
http://api.brain-map.org//api/v2/well_known_file_download/1026123653
http://api.brain-map.org//api/v2/well_known_file_download/1026123655
http://api.brain-map.org//api/v2/well_known_file_download/1026123661
http://api.brain-map.org//api/v2/well_known_file_download/1026124481
http://api.brain-map.org//api/v2/well_known_file_download/1026124479
http://api.brain-map.org//api/v2/well_known_file_download/1026124471
http://api.brain-map.org//api/v2/well_known_file_download/1026124473
http://api.brain-map.org//api/v2/well_known_file_download/1026124475
http://api.brain-map.org//api/v2/well_known_file_download/1026124477
http://api.brain-map.org//api/v2/well_known_file_download/1026124761
http://api.brain-map.org//api/v2/well_known_file_download/1026124765
http://api.brain-map.org//api/v2/well_known_file_download/1026124763
http://api.brain-map.org//api/v2/well_known_file_download/1026124767
http://api.brain-map.org//api/v2/well_known_file_download/1026124769
http://api.brain-map.org//api/v2/well_known_file_download/1026124771
http://api.brain-map.org//api/v2/well_known_file_download/1026124097
http://api.brain-map.org//api/v2/well_known_file_download/1026124095
http://api.brain-map.org//api/v2/well_known_file_download/1026124093
http://api.brain-map.org//api/v2/well_known_file_download/1026124088
http://api.brain-map.org//api/v2/well_known_file_download/1026124091
http://api.brain-map.org//api/v2/well_known_file_download/1026123830
http://api.brain-map.org//api/v2/well_known_file_download/1026123828
http://api.brain-map.org//api/v2/well_known_file_download/1026123834
http://api.brain-map.org//api/v2/well_known_file_download/1026123832
http://api.brain-map.org//api/v2/well_known_file_download/1026123826
http://api.brain-map.org//api/v2/well_known_file_download/1026123635
http://api.brain-map.org//api/v2/well_known_file_download/1026123631
http://api.brain-map.org//api/v2/well_known_file_download/1026123629
http://api.brain-map.org//api/v2/well_known_file_download/1026123633
http://api.brain-map.org//api/v2/well_known_file_download/1026123627
http://api.brain-map.org//api/v2/well_known_file_download/1026123791
http://api.brain-map.org//api/v2/well_known_file_download/1026123793
http://api.brain-map.org//api/v2/well_known_file_download/1026123795
http://api.brain-map.org//api/v2/well_known_file_download/1026123789
http://api.brain-map.org//api/v2/well_known_file_download/1026123797
file for probe 832810584 not found
http://api.brain-map.org//api/v2/well_known_file_download/1026124842
http://api.brain-map.org//api/v2/well_known_file_download/1026124846
http://api.brain-map.org//api/v2/well_known_file_download/1026124844
http://api.brain-map.org//api/v2/well_known_file_download/1026124848
http://api.brain-map.org//api/v2/well_known_file_download/1026124850
http://api.brain-map.org//api/v2/well_known_file_download/1026124374
http://api.brain-map.org//api/v2/well_known_file_download/1026124370
http://api.brain-map.org//api/v2/well_known_file_download/1026124378
http://api.brain-map.org//api/v2/well_known_file_download/1026124376
http://api.brain-map.org//api/v2/well_known_file_download/1026124372
http://api.brain-map.org//api/v2/well_known_file_download/1026123521
http://api.brain-map.org//api/v2/well_known_file_download/1026123524
http://api.brain-map.org//api/v2/well_known_file_download/1026123526
http://api.brain-map.org//api/v2/well_known_file_download/1026123528
http://api.brain-map.org//api/v2/well_known_file_download/1026123530
http://api.brain-map.org//api/v2/well_known_file_download/1026124428
http://api.brain-map.org//api/v2/well_known_file_download/1026124431
http://api.brain-map.org//api/v2/well_known_file_download/1026124426
http://api.brain-map.org//api/v2/well_known_file_download/1026124424
http://api.brain-map.org//api/v2/well_known_file_download/1026124437
http://api.brain-map.org//api/v2/well_known_file_download/1026124443
http://api.brain-map.org//api/v2/well_known_file_download/1026123729
http://api.brain-map.org//api/v2/well_known_file_download/1026123733
http://api.brain-map.org//api/v2/well_known_file_download/1026123731
http://api.brain-map.org//api/v2/well_known_file_download/1026123727
http://api.brain-map.org//api/v2/well_known_file_download/1026123738
http://api.brain-map.org//api/v2/well_known_file_download/1026123735
file for probe 846401828 not found
file for probe 846401830 not found
file for probe 846401834 not found
file for probe 846401838 not found
file for probe 846401841 not found
http://api.brain-map.org//api/v2/well_known_file_download/1026123956
http://api.brain-map.org//api/v2/well_known_file_download/1026123951
http://api.brain-map.org//api/v2/well_known_file_download/1026123953
http://api.brain-map.org//api/v2/well_known_file_download/1026123949
http://api.brain-map.org//api/v2/well_known_file_download/1026123945
http://api.brain-map.org//api/v2/well_known_file_download/1026123947
Invalid Time Intervals#
On some occasions there were problems with data acquisition for a brief period of time. These problems didn’t necessitate failing the entire experiment, but they did invalidate the data during those times. Depending on your analysis goals, you might need to work around these invalid times. Because these problems occur rarely, built-in methods do not filter them out. If a probe you are analyzing has such invalid times, one method for addressing this is to filter out trials from the stimulus table with a start_time or stop_time that overlaps with any of the invalid intervals.
When we access the spike_times of a session with invalid time intervals for the first time, we get a warning about it:
# Example session with invalid times
session_id = 732592105
session = cache.get_session_data(session_id=session_id)
session.spike_times;
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'hdmf-common' version 1.1.3 because version 1.8.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.2.2 because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'hdmf-common' version 1.1.3 because version 1.8.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.2.2 because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/allensdk/brain_observatory/ecephys/ecephys_session.py:1371: UserWarning: Session includes invalid time intervals that could be accessed with the attribute 'invalid_times',Spikes within these intervals are invalid and may need to be excluded from the analysis.
warnings.warn("Session includes invalid time intervals that could "
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'hdmf-common' version 1.1.3 because version 1.8.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.2.2 because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'hdmf-common' version 1.1.3 because version 1.8.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.2.2 because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'hdmf-common' version 1.1.3 because version 1.8.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.2.2 because version 2.7.0 is already loaded.
return func(args[0], **pargs)
Notice the warning:
UserWarning: Session includes invalid time intervals that could be accessed with the attribute ‘invalid_times’,Spikes within these intervals are invalid and may need to be excluded from the analysis.
You can determine which time intervals are invalid using session.get_invalid_times() which returns a DataFrame with the start and stop time of intervals of invalid data and the associated probes. You can always use this to check whether a session has invalid times — if it doesn’t, the returned DataFrame will be empty — but if you don’t see a warning, it is free of invalid intervals.
session.get_invalid_times()
| start_time | stop_time | tags | |
|---|---|---|---|
| id | |||
| 0 | 882.52 | 962.56 | [EcephysProbe, 733744653, probeE] |
| 1 | 4148.40 | 4214.91 | [EcephysProbe, 733744653, probeE] |
| 2 | 6841.77 | 6923.46 | [EcephysProbe, 733744653, probeE] |
| 3 | 8843.23 | 8911.38 | [EcephysProbe, 733744653, probeE] |
The tags give us information about what caused the invalid times. In this case, they were caused by a malfunctioning probe. We can examine the data on that probe and see that during the invalid timespans, the data are NaN.
bad_probe_id = 733744653
timespan = slice(882.517, 962.562)
# Get the Local Field Potentials from the temporarily malfunctioning probe
lfp = session.get_lfp(bad_probe_id)
lfp.sel(time=timespan).to_pandas()
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'hdmf-common' version 1.1.3 because version 1.8.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.2.2 because version 2.7.0 is already loaded.
return func(args[0], **pargs)
| channel | 850233713 | 850233721 | 850233729 | 850233737 | 850233745 | 850233753 | 850233761 | 850233769 | 850233777 | 850233785 | ... | 850234345 | 850234353 | 850234361 | 850234369 | 850234377 | 850234385 | 850234393 | 850234401 | 850234409 | 850234417 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| time | |||||||||||||||||||||
| 882.517637 | 0.000149 | 0.000309 | 0.000317 | 0.000127 | 0.000171 | 0.000213 | 0.000124 | 0.000222 | 0.000262 | 0.000153 | ... | -0.000129 | 0.000004 | 0.000031 | 0.000068 | 0.000013 | -0.000029 | 0.000211 | -0.000026 | -0.000004 | -0.000033 |
| 882.518437 | 0.000163 | 0.000319 | 0.000331 | 0.000144 | 0.000127 | 0.000190 | 0.000120 | 0.000235 | 0.000290 | 0.000164 | ... | -0.000133 | 0.000003 | 0.000036 | 0.000052 | 0.000013 | -0.000037 | 0.000206 | -0.000043 | -0.000003 | -0.000049 |
| 882.519237 | 0.000194 | 0.000324 | 0.000369 | 0.000183 | 0.000171 | 0.000217 | 0.000150 | 0.000254 | 0.000317 | 0.000200 | ... | -0.000127 | -0.000011 | 0.000032 | 0.000064 | 0.000022 | -0.000017 | 0.000222 | -0.000028 | 0.000011 | -0.000026 |
| 882.520037 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 882.520837 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 962.558155 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 962.558955 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 962.559755 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 962.560555 | -0.000083 | -0.000229 | -0.000300 | -0.000129 | -0.000078 | -0.000158 | -0.000077 | -0.000179 | -0.000233 | -0.000129 | ... | 0.000081 | -0.000055 | -0.000074 | -0.000076 | -0.000015 | 0.000015 | -0.000188 | 0.000027 | 0.000025 | 0.000038 |
| 962.561355 | -0.000091 | -0.000194 | -0.000278 | -0.000136 | -0.000049 | -0.000148 | -0.000057 | -0.000150 | -0.000214 | -0.000114 | ... | 0.000054 | -0.000079 | -0.000085 | -0.000046 | -0.000011 | 0.000011 | -0.000188 | 0.000029 | 0.000028 | 0.000046 |
1253 rows × 89 columns
For more information on why there were issues with data acquisition in these instances, see the “Invalid intervals” section of our whitepaper.
