Visual Coding — Neuropixels#
The Visual Coding – Neuropixels project uses high-density extracellular electrophysiology (Ecephys) probes to record spikes from a wide variety of regions in the mouse brain. Our experiments are designed to study the activity of the visual cortex and thalamus in the context of passive visual stimulation as is described in Siegle et al. [2021], but these data can be used to address a wide variety of topics.
Spike-sorted data and metadata are available via the AllenSDK as Neurodata Without Borders (NWB) files [Teeters et al., 2015]. However, if you’re using the AllenSDK to interact with the data, no knowledge of the NWB data format is required.
Getting started#
To jump right in, check out the quick start guide, which will show you how to download the data, align spikes to a visual stimulus, and decode natural images from neural activity patterns. For a quick summary of experimental design and data access, see the Cheat sheet for visual coding Neuropixels.
If you would like more example code, the full example notebook covers all of the ways to access data for each experiment.
Additional tutorials are available on the following topics:
For detailed information about the experimental design, data acquisition, and informatics methods, please refer to our technical whitepaper. AllenSDK API documentation is available here.
Note
A note on terminology: Throughout the SDK, we refer to neurons as units, because we cannot guarantee that all the spikes assigned to one unit actually originate from a single cell. Unlike in two-photon imaging, where you can visualize each neuron throughout the entire experiment, with electrophysiology we can only “see” a neuron when it fires a spike. If a neuron moves relative to the probe, or if the neuron is far away from the probe, some of its spikes may get mixed together with those from other neurons. Because of this inherent ambiguity, we provide a variety of quality metrics to allow you to find the right units for your analysis. Even highly contaminated units contain potentially valuable information about brain states, so they are still included within the dataset. However, certain types of analysis require more stringent quality thresholds to ensure that all of the included units are well isolated from their neighbors.
Data processing#
See the section on Neuropixels data processing.
Visual stimulus sets#
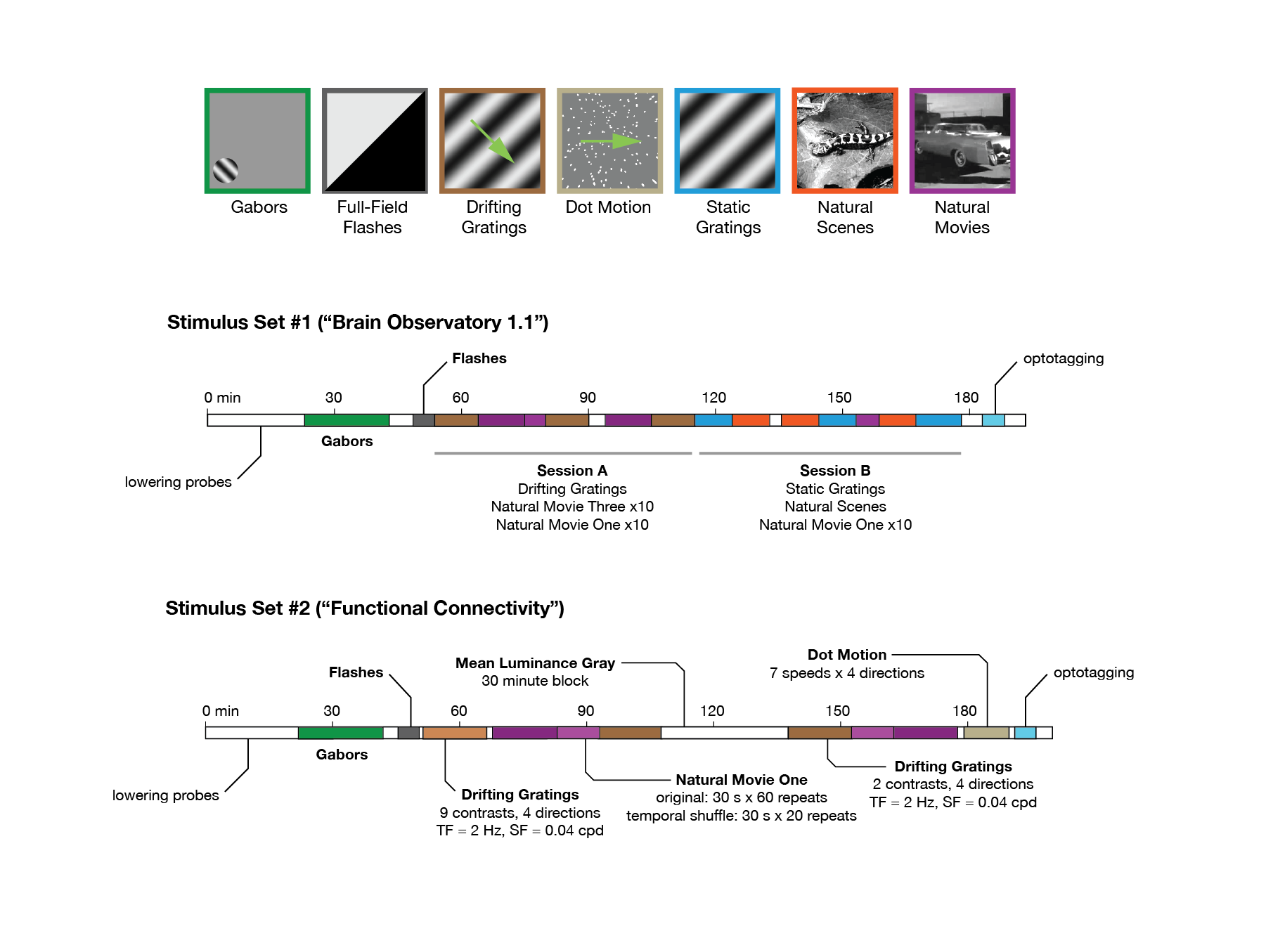
Fig. 17 Neuropixels visual stimulus sets#
A central aim of the Visual Coding – Neuropixels project is to measure the impact of visual stimuli on neurons throughout the mouse visual system. To that end, all mice viewed one of two possible stimulus sets, known as Brain Observatory 1.1 or Functional Connectivity. Both stimulus sets began with a Gabor stimulus flashed at 81 different locations on the screen, used to map receptive fields of visually responsive units. Next, the mice were shown brief flashes of light or dark, to measure the temporal dynamics of the visual response.
The remainder of the visual stimulus set either consisted of the same stimuli shown in the two-photon experiments (Brain Observatory 1.1), or a subset of those stimuli shown with a higher number of repeats. We also added a dot motion stimulus, to allow us to measure the speed tuning of units across the mouse visual system.
Quality metrics#
Every NWB file includes a table of quality metrics, which can be used to assess
the completeness, contamination, and stability of units in the recording. By
default, we won’t show you units below a pre-determined quality threshold; we
hide any units that are not present for the whole session
(presence_ratio < 0.95), that include many contaminating spikes
(isi_violations > 0.5), or are likely missing a large fraction of spikes
(amplitude_cutoff > 0.1). However, even contaminated or incomplete units contain
information about brain states, and may be of interest to analyze. Therefore,
the complete units table can be accessed via special flags in the AllenSDK.
In general, we do not make a distinction between ‘single-unit’ and ‘multi-unit’
activity. There is no obvious place to draw a boundary in the overall
distributions of quality metrics, and setting a strict cutoff (e.g.
isi_violations = 0) will remove a lot of potentially valuable data. We prefer to
leave it up to the end user to decide what level of contamination is tolerable.
But that means you need to be aware that different units will have different
levels of cleanliness.
It should also be noted that all of these metrics assume that the spike waveform is stable throughout the experiment. Given that the probe drifts, on average, about 40 μm over the course of the ~3 hour recordings, this assumption is almost never valid. The resulting changes in waveform shape can cause a unit’s quality to fluctuate. If you’re unsure about a unit’s quality, it can be helpful to plot its spike amplitudes over time. This can make it obvious if it’s drifting below threshold, or if it contains spikes from multiple neurons.
Documentation on the various quality metrics can be found in the ecephys_spike_sorting repository.
For a detailed discussion of the appropriate way to apply each of these metrics, please check out the unit quality metric tutorial.
Precomputed stimulus metrics#
Tables of precomputed metrics are available for download to support population
analysis and filtering. The table below describes all of the available metrics.
The get_unit_analysis_metrics() method will load this table as a
pandas DataFrame.
drifting gratings#
Metric |
Field Name |
|---|---|
preferred orientation |
|
preferred temporal frequency |
|
global orientation selectivity |
|
global direction selectivity |
|
running modulation |
|
running modulation p-value |
|
firing rate |
|
fano factor |
|
modulation index |
|
f1/f0 |
|
lifetime sparseness |
|
c50 (contrast tuning stimulus) |
|
static gratings#
Metric |
Field Name |
|---|---|
preferred orientation |
|
preferred spatial frequency |
|
preferred phase |
|
global orientation selectivity |
|
running modulation |
|
running modulation p-value |
|
firing rate |
|
fano factor |
|
lifetime sparseness |
|
natural scenes#
Metric |
Field Name |
|---|---|
preferred image index |
|
image selectivity |
|
running modulation |
|
running modulation p-value |
|
firing rate |
|
fano factor |
|
lifetime sparseness |
|
dot motion#
Metric |
Field Name |
|---|---|
preferred speed |
|
preferred direction |
|
running modulation |
|
running modulation p-value |
|
firing rate |
|
fano factor |
|
lifetime sparseness |
|
full-field flashes#
Metric |
Field Name |
|---|---|
on/off ratio |
|
running modulation |
|
running modulation p-value |
|
firing rate |
|
fano factor |
|
lifetime sparseness |
|
gabors#
Metric |
Field Name |
|---|---|
RF area |
|
RF elevation |
|
RF azimuth |
|
RF p-value |
|
running modulation |
|
running modulation p-value |
|
firing rate |
|
fano factor |
|
lifetime sparseness |
|
AllenSDK 2.0 and backwards data compatibility#
AllenSDK version 2.0 marks a major update to released Visual Coding Neuropixels datasets. Due to newer versions of pynwb/hdmf having issues reading previously released Visual Coding Neuropixels NWB files and due to the significant reorganization of updated NWB file contents, this release contains breaking changes that necessitate a major version revision. NWB files released prior to 6/11/2020 are not guaranteed to work with the 2.0.0 version of AllenSDK. If you cannot or choose not to re-download the updated NWB files, you can continue using a prior version of AllenSDK (< 2.0.0) to access them. However, no further features or bugfixes for AllenSDK (< 2.0.0) are planned. Data released for other projects (Cell Types, Mouse Connectivity, etc.) are NOT affected and will NOT need to be re-downloaded.
When using the Visual Coding EcephysProjectCache from this updated AllenSDK
version, if a ManifestError is encountered, this indicates that previously
downloaded cached data files need to be removed and re-downloaded. The location
these files as well as manifest are user defined and are set when instantiating
an EcephysProjectCache.
