Neuroglancer#
Note
Neuroglancer works best in Chrome and Firefox but does not always work as expected in Safari.
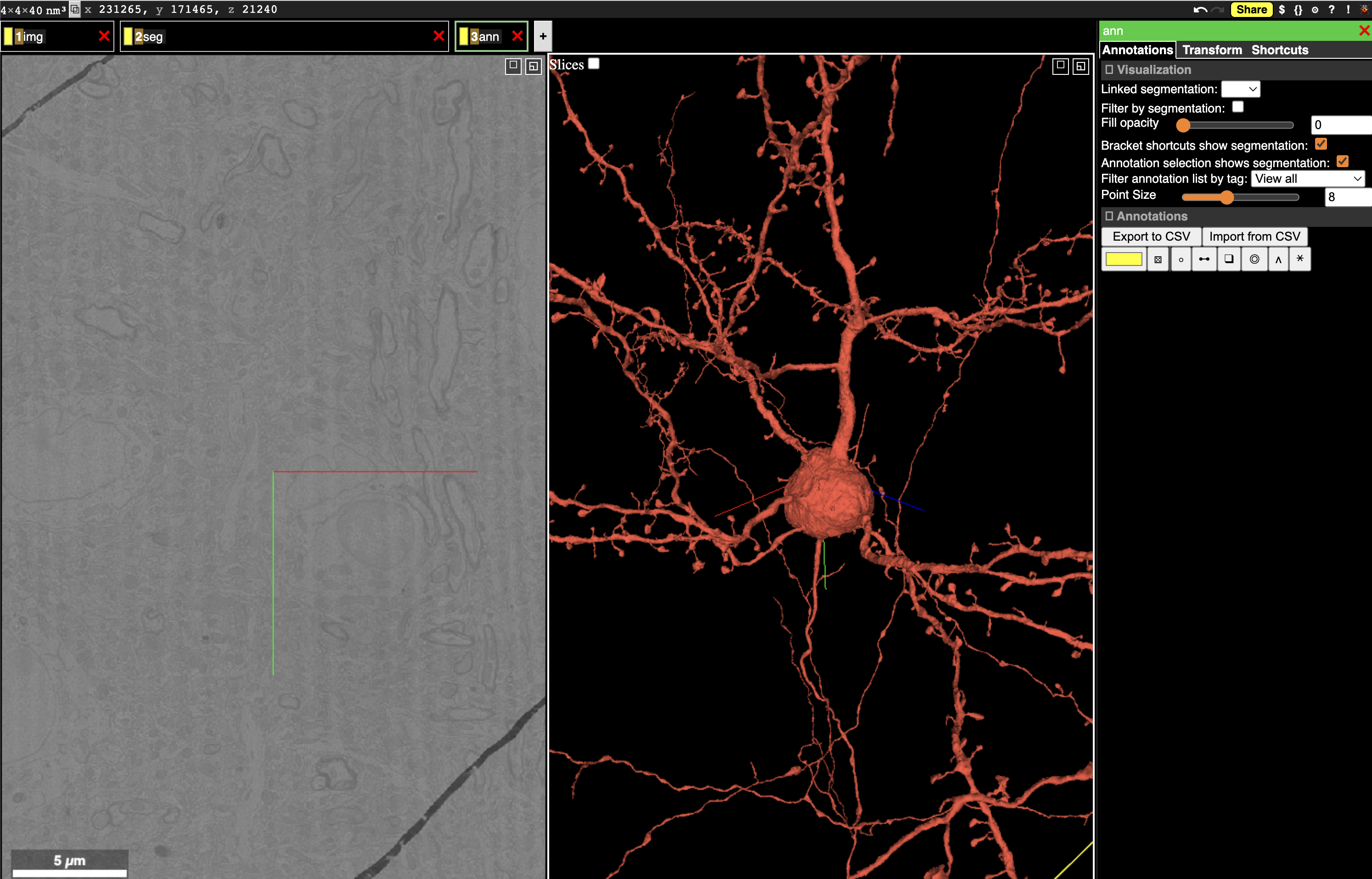
Fig. 28 Neuroglancer can show multiple layers of data, including imagery, segmentation, and more.#
Neuroglancer is a WebGL-based viewer developed by Jeremy Maitin-Shephard at the Google Connectomics team to visualize very large volumetric data, designed in large part for connectomics. We often use Neuroglancer to quickly explore data, visualize results in context, and share data.
To view MICrONS data in neuroglancer: MICrONS Neuroglancer
To view V1DD data in neuroglancer: V1DD Neuroglancer
Note that you will need to authenticate with the same Google-associated account that you use to set up CAVEclient.
For animated data access examples, see MICrONS Explorer Neuroglancer introduction
Dataset specificity#
The examples below are written with the MICrONS dataset. To access the V1DD dataset change the datastack from minnie65_public to v1dd_public
Interface Basics#
The Neuroglancer interface is divided into panels. In the default view, one panel shows the imagery in the X/Y plane (left), one shows a 3d view centered at the same location, and the narrow third panel provides information about the specific layer.
Note that at the center of the each panel is a collection of axis-aligned red, blue and, green lines. The intersection and direction of each of these lines is consistent across all panels.
Along the top left of the view, you can see tabs with different names. Neuroglancer organizes data into layers, where each layer tells Neuroglancer about a different aspect of the data. The default view has three layers:
imgdescribes how to render imagery.segdescribes how to render segmentation and meshes.annis a manual annotation layer, allowing the user to add annotations to the data.
You can switch between layers by right clicking on the layer tab. You will see the panel at the right change to provide controls for each layer as you click it.
The collection of all layers, the user view, and all annotations is stored as a JSON object called the state.
The basic controls for navigation are:
left click/dragslides the imagery in X/Y and rotates the 3d view.scroll wheel up/downmoves the imagery in Z. (track pad two finger scroll; keypad , or . )right clickjumps the 3d view to the clicked location in either the imagery or on a segmented object.double left clickselects a segmentation and loads its mesh into the 3d view. Double clicking on a selected neuron deselects it.control-scrollzooms the view under the cursor in or out. (track pad two finger expand/contract)zsnaps the view to the closest right angle.
You can paste a position into Neuroglancer by clicking the x, y, z coordinate in the upper left corner and pasting a space or comma-separated list of numbers and hitting enter. Note that Neuroglancer always works in voxel units, and you can see the resolution of the voxels in the extreme upper left corner.
Selecting objects#
The most direct way to select a neuron is to double click in the imagery to select the object under your cursor. This will load all the voxels associated with that object and also display its mesh in the 3d view.
To see the list of selected objects, you can select the segmentation tab (right click on the seg tab).
Underneath the list of options, there is a list of selected root ids and the color assigned to them in the view.
You can change colors of all neurons randomly by pressing l or individually change colors as desired.
In addition, you can press the checkbox to hide a selected object while keeping it in the list, or deselect it by clicking on the number itself. You can also copy a root id by pressing the clipboard icon next to its number, or copy all selected root ids by pressing the clipboard icon above the list.
This selection list also allows you to select objects by pasting one or more root ids into the text box at the top of the list and pressing enter.
Annotations#
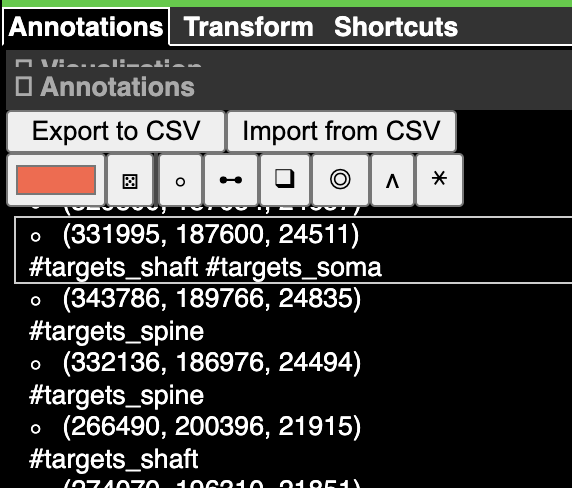
Annotations are stored in an annotation layer.
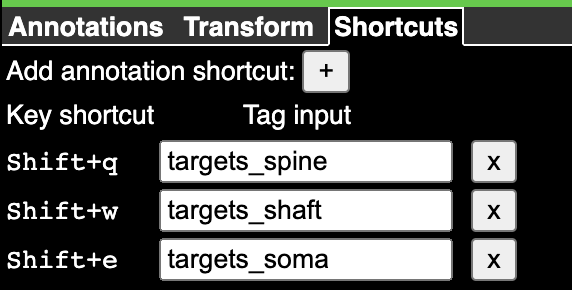
The default state has an annotation layer called ann, but you can always add new annotation layers by command-clicking the + button to the right of the layer tabs.
To create an annotation, select the layer (right click on the tab), and then click the icon representing the type of annotation you want to use. The most basic annotation is a point, which is the icon to the left of the list. The icon will change to having a green background when selected.
Now if you control-click in either the imagery or the 3d view, you will create a point annotation at the clicked location. The annotation will appear in the list to the right, with its coordinate (in voxels, not nanometers) displayed. Clicking any annotation in the list will jump to that annotation in 3d space. Each annotation layer can have one color, which you can change with the color picker to the left of the annotation list.
Saving and sharing states#
Like many other websites that require logins, you cannot simply send your URL ot another person to have them see the view.
Instead, to save the current state and make it available to yourself or others in the future, you need to save the state with the Share button at the top right corner.
This will then give you a URL that you can copy and share with others or paste yourself.
A typical sharing URL looks like the following:
https://spelunker.cave-explorer.org/#!middleauth+https://global.daf-apis.com/nglstate/api/v1/4658335189041152
The first part is the URL for the Neuroglancer viewer, while the part after the ?json_url= is a URL that points to a JSON file that contains the state.
The number at the end of the URL is used to uniquely identify the state and can be used programmatically to retrieve information.
Warning
If a URL contains ?local_id= instead of ?json_url, that means that it cannot be viewed by anyone else or even in another browser on your own computer.
Programmatic Interaction with Neuroglancer States#
Important
Before using any programmatic access to the data, you first need to set up your CAVEclient token.
Visualizing data in Neuroglancer is one of the easiest ways to explore it in its full context.
The python package nglui was made to make it easy to generate Neuroglancer states from data, particularly pandas dataframes, in a progammatic manner.
The package can be installed with pip install nglui.
Note
The nglui package interacts prominently with caveclient and annotations queried from the database.
See the section on querying the database to learn more.
Parsing Neuroglancer states#
The nglui.parser module offers a number of tools to get information about neuroglancer states out of the JSON format that neuroglancer uses. The recommended approach here is to pass a dictionary representation of the JSON object the StateParser class and build various kinds of dataframes from it.
The simplest way to parse the annotations in a Neuroglancer state is to first save the state using the Share button, and then copy the state id (the last number in the URL).
Important
This requires nglui version >= 3.5.0 and <4.0.0
from caveclient import CAVEclient
from nglui import parser
client = CAVEclient('minnie65_public')
state_json = client.state.get_state_json(6107390807113728)
state_parser = parser.StateParser(state_json)
You can now access different aspects of the state through this state_parser object.
Layer Data#
For example, to get a list of all layers and their core info, you can use the layer_dataframe method
state_parser.layer_dataframe()
will give you a table with a row for each layer and columns for layer name, type, source, and whether the layer is archived (i.e. visible) or not.
You can also get a list of all selected segments with the selection_dataframe method.
state_parser.selection_dataframe()
will give you a dataframe where each row is a selected segment, and columns show layer name, segment id, and whether or not the segment is visible.
Annotation Data#
Finally, you can get a list of all annotations withthe annotation_dataframe method. h
state_parser.annotation_dataframe()
will give you a dataframe where each row is an annotation, and columns show layer name, points locations, annotation type, annotation id, linked segmentations, tags, etc.
If you are using tags, the expand_tags=True argument will create a column for every tag and assign a boolean value to the row based on whether the tag is present in the annotation.
Another option that is sometimes useful is split_points=True, which will create a separate column for each x, y, or z coordinate in the annotation.od.d..
Generating Neuroglancer States from Data#
The nglui.statebuilder package is used to build Neuroglancer states that express arbitrary data.
The general pattern is that one makes a “StateBuilder” object that has rules for how to build a Neuroglancer state layer by layer, including selecting certain neurons, and populate layers of annotations.
You then pass a DataFrame to the StateBuiler, and the rules tell it how to render the DataFrame into a Neuroglancer link.
The same set of rules can be used on similar dataframes but with different data, such as synapses from different neurons.
To understand the detailed use of the package, please see the the NGLUI documentation (and mind the version differences).
However, a number of basic helper functions allow nglui to be used for common functions in just a few lines.
For example, to generate a Neuroglancer state that shows a neuron and its synaptic inputs and outputs, we can use the make_neuron_neuroglancer_link helper function.
from caveclient import CAVEclient
from nglui.statebuilder import helpers
client = CAVEclient('minnie65_public')
helpers.make_neuron_neuroglancer_link(
client,
root_ids=864691135441799752,
show_inputs=True,
show_outputs=True,
target_site='cave-explorer',
)
(<nglui.statebuilder.statebuilder.ChainedStateBuilder at 0x7fe98359cdc0>,
[ root_id
0 864691135441799752,
id created superceded_id valid \
148 187770131 2020-11-04 07:36:45.318243+00:00 NaN t
181 173033456 2020-11-04 07:36:40.962629+00:00 NaN t
185 180581870 2020-11-04 07:36:39.970777+00:00 NaN t
706 165299728 2020-11-04 10:15:19.696268+00:00 NaN t
760 194395546 2020-11-04 07:36:39.970777+00:00 NaN t
... ... ... ... ...
4394 163655586 2020-11-04 08:04:15.811361+00:00 NaN t
2434 192764780 2020-11-04 08:37:14.648836+00:00 NaN t
2843 194462083 2020-11-04 11:47:21.612406+00:00 NaN t
1622 165048795 2020-11-04 14:51:00.402773+00:00 NaN t
398 207066737 2020-11-04 08:40:11.026602+00:00 NaN t
pre_pt_position_x pre_pt_position_y pre_pt_position_z \
148 192446 119188 20874
181 187016 115800 21592
185 191728 119586 20929
706 183288 120324 19046
760 197002 119200 21290
... ... ... ...
4394 180908 105584 23021
2434 199496 110902 22560
2843 198824 103000 21123
1622 183558 104924 21713
398 204946 114046 19339
post_pt_position_x post_pt_position_y post_pt_position_z \
148 192498 119260 20887
181 186944 115842 21583
185 191820 119510 20927
706 183390 120246 19047
760 196906 119330 21295
... ... ... ...
4394 180918 105622 23031
2434 199538 110820 22558
2843 198780 102884 21131
1622 183576 104896 21720
398 204922 114008 19338
ctr_pt_position_x ctr_pt_position_y ctr_pt_position_z size \
148 192419 119264 20880 2228
181 187006 115770 21584 5096
185 191780 119582 20929 2684
706 183354 120302 19043 3348
760 196972 119256 21290 2632
... ... ... ... ...
4394 180923 105549 23019 9432
2434 199524 110864 22557 2324
2843 198816 102938 21126 2400
1622 183590 104922 21714 3900
398 204954 114014 19341 2908
pre_pt_supervoxel_id pre_pt_root_id post_pt_supervoxel_id \
148 91205727213274611 864691136010404012 91205727213292378
181 90501558869035020 864691136010404012 90501558869029887
185 91135427188633134 864691136010404012 91135427188640584
706 89939227055371613 864691136010404012 90009595799564883
760 91839045977868681 864691136010404012 91839045977876607
... ... ... ...
4394 89655759684092822 864691131915395450 89655759684111608
2434 92189790388170110 864691131889640562 92189790388169159
2843 92118321931148216 864691131872674215 92118321931159175
1622 90007534550924139 864691131822625017 90007534550931742
398 92964258421597883 864691131794106944 92964258421600601
post_pt_root_id
148 864691135441799752
181 864691135441799752
185 864691135441799752
706 864691135441799752
760 864691135441799752
... ...
4394 864691135441799752
2434 864691135441799752
2843 864691135441799752
1622 864691135441799752
398 864691135441799752
[5681 rows x 18 columns],
id created superceded_id valid \
346 188722054 2020-11-04 09:25:30.539182+00:00 NaN t
690 162924603 2020-11-04 07:53:27.186324+00:00 NaN t
1496 162925312 2020-11-04 07:53:40.155613+00:00 NaN t
1633 151730182 2020-11-04 09:39:15.637331+00:00 NaN t
1880 151623255 2020-11-04 09:25:21.569113+00:00 NaN t
... ... ... ... ...
888 343810565 2020-11-04 14:11:24.798884+00:00 NaN t
178 173134723 2020-11-04 08:05:35.030894+00:00 NaN t
2173 151242210 2020-11-04 11:30:20.588254+00:00 NaN t
584 191679382 2020-11-04 11:18:38.997022+00:00 NaN t
1135 189709681 2020-11-04 13:37:56.862784+00:00 NaN t
pre_pt_position_x pre_pt_position_y pre_pt_position_z \
346 192024 157924 21904
690 178168 232030 24446
1496 179160 232006 24380
1633 174736 101966 22287
1880 172330 99914 22442
... ... ... ...
888 287542 91132 20636
178 186522 121764 20250
2173 171458 98266 23004
584 196276 115870 19738
1135 192476 195542 20379
post_pt_position_x post_pt_position_y post_pt_position_z \
346 192082 157964 21903
690 178168 232072 24455
1496 179154 231910 24379
1633 174738 101932 22276
1880 172268 99934 22438
... ... ... ...
888 287438 91160 20648
178 186594 121722 20262
2173 171482 98312 22999
584 196228 115776 19737
1135 192454 195502 20373
ctr_pt_position_x ctr_pt_position_y ctr_pt_position_z size \
346 192036 157910 21903 3444
690 178156 232137 24449 21364
1496 179182 231938 24382 9788
1633 174758 101982 22281 6060
1880 172254 99910 22444 16348
... ... ... ... ...
888 287480 91184 20643 12084
178 186563 121727 20258 3460
2173 171448 98312 22999 2000
584 196254 115798 19739 2376
1135 192426 195532 20374 3264
pre_pt_supervoxel_id pre_pt_root_id post_pt_supervoxel_id \
346 91140581283589200 864691135441799752 91210950027716191
690 89250521130992077 864691135441799752 89250521131006598
1496 89391258619218995 864691135441799752 89391258619228831
1633 88810853650201222 864691135441799752 88810853650184703
1880 88458735051562945 864691135441799752 88458735051562962
... ... ... ...
888 104290534058860946 864691135441799752 104290534058874947
178 90432014557495008 864691135441799752 90432014557505248
2173 88317791471923132 864691135441799752 88317791471919946
584 91768195996011195 864691135441799752 91768195996012041
1135 91215966348199249 864691135441799752 91215966348192937
post_pt_root_id
346 864691135517376211
690 864691135517376211
1496 864691135517376211
1633 864691135517376211
1880 864691135517376211
... ...
888 864691131857510409
178 864691131757970238
2173 864691131746523286
584 864691131737138899
1135 864691131599924730
[2231 rows x 18 columns]])
The main helper functions are:
make_neuron_neuroglancer_link- Shows one or more neurons and, optionally, synaptic inputs and/or outputs.make_synapse_neuroglancer_link- Using a pre-downloaded synapse table, make a link that shows the synapse and the listed synaptic partners.make_point_statebuilder- Generate a statebuilder that to map a dataframe containing points (by default, formatted like a cell types table) to a Neuroglancer link.
In all cases, please look at the docstrings for more information on how to use the functions.