Full example of accessing Visual Coding Neuropixels data#
At the Allen Institute for Brain Science we carry out in vivo extracellular electrophysiology (ecephys) experiments in awake animals using high-density Neuropixels probes. The data from these experiments are organized into sessions, where each session is a distinct continuous recording period. During a session we collect:
spike times and characteristics (such as mean waveforms) from up to 6 neuropixels probes
local field potentials
behavioral data, such as running speed and eye position
visual stimuli which were presented during the session
cell-type specific optogenetic stimuli that were applied during the session
The AllenSDK contains code for accessing across-session (project-level) metadata
as well as code for accessing detailed within-session data. The standard
workflow is to use project-level tools, such as EcephysProjectCache to
identify and access sessions of interest, then delve into those sessions’ data
using EcephysSession.
Project-level#
The EcephysProjectCache class in
allensdk.brain_observatory.ecephys.ecephys_project_cache accesses and stores
data pertaining to many sessions. You can use this class to run queries that
span all collected sessions and to download data for individual sessions.
Session-level#
The EcephysSession class in
allensdk.brain_observatory.ecephys.ecephys_session provides an interface to
all of the data for a single session, aligned to a common clock. This notebook
will show you how to use the EcephysSession class to extract these data.
# first we need a bit of import boilerplate
import os
import numpy as np
import xarray as xr
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
from scipy.ndimage.filters import gaussian_filter
from pathlib import Path
import json
from IPython.display import display
from PIL import Image
from allensdk.brain_observatory.ecephys.ecephys_project_cache import EcephysProjectCache
from allensdk.brain_observatory.ecephys.ecephys_session import (
EcephysSession,
removed_unused_stimulus_presentation_columns
)
from allensdk.brain_observatory.ecephys.visualization import plot_mean_waveforms, plot_spike_counts, raster_plot
from allensdk.brain_observatory.visualization import plot_running_speed
# tell pandas to show all columns when we display a DataFrame
pd.set_option("display.max_columns", None)
/tmp/ipykernel_9685/2939167369.py:9: DeprecationWarning: Please use `gaussian_filter` from the `scipy.ndimage` namespace, the `scipy.ndimage.filters` namespace is deprecated.
from scipy.ndimage.filters import gaussian_filter
/opt/envs/allensdk/lib/python3.10/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
Obtaining an EcephysProjectCache#
In order to create an EcephysProjectCache object, you need to specify two
things:
A remote source for the object to fetch data from. We will instantiate our cache using
EcephysProjectCache.from_warehouse()to point the cache at the Allen Institute’s public web API.A path to a manifest json, which designates filesystem locations for downloaded data. The cache will try to read data from these locations before going to download those data from its remote source, preventing repeated downloads.
output_dir = '/root/capsule/data/allen-brain-observatory/visual-coding-neuropixels/ecephys-cache/'
manifest_path = os.path.join(output_dir, "manifest.json")
resources_dir = Path('/opt/databook/databook/resources')
DOWNLOAD_LFP = False
# Example cache directory path, it determines where downloaded data will be stored
cache = EcephysProjectCache.from_warehouse(manifest=manifest_path)
Querying across sessions#
Using your EcephysProjectCache, you can download a table listing metadata for
all sessions.
cache.get_session_table().head()
| published_at | specimen_id | session_type | age_in_days | sex | full_genotype | unit_count | channel_count | probe_count | ecephys_structure_acronyms | |
|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||
| 715093703 | 2019-10-03T00:00:00Z | 699733581 | brain_observatory_1.1 | 118.0 | M | Sst-IRES-Cre/wt;Ai32(RCL-ChR2(H134R)_EYFP)/wt | 884 | 2219 | 6 | [CA1, VISrl, nan, PO, LP, LGd, CA3, DG, VISl, ... |
| 719161530 | 2019-10-03T00:00:00Z | 703279284 | brain_observatory_1.1 | 122.0 | M | Sst-IRES-Cre/wt;Ai32(RCL-ChR2(H134R)_EYFP)/wt | 755 | 2214 | 6 | [TH, Eth, APN, POL, LP, DG, CA1, VISpm, nan, N... |
| 721123822 | 2019-10-03T00:00:00Z | 707296982 | brain_observatory_1.1 | 125.0 | M | Pvalb-IRES-Cre/wt;Ai32(RCL-ChR2(H134R)_EYFP)/wt | 444 | 2229 | 6 | [MB, SCig, PPT, NOT, DG, CA1, VISam, nan, LP, ... |
| 732592105 | 2019-10-03T00:00:00Z | 717038288 | brain_observatory_1.1 | 100.0 | M | wt/wt | 824 | 1847 | 5 | [grey, VISpm, nan, VISp, VISl, VISal, VISrl] |
| 737581020 | 2019-10-03T00:00:00Z | 718643567 | brain_observatory_1.1 | 108.0 | M | wt/wt | 568 | 2218 | 6 | [grey, VISmma, nan, VISpm, VISp, VISl, VISrl] |
Querying across probes#
… or for all probes
cache.get_probes().head()
| ecephys_session_id | lfp_sampling_rate | name | phase | sampling_rate | has_lfp_data | unit_count | channel_count | ecephys_structure_acronyms | |
|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||
| 729445648 | 719161530 | 1249.998642 | probeA | 3a | 29999.967418 | True | 87 | 374 | [APN, LP, MB, DG, CA1, VISam, nan] |
| 729445650 | 719161530 | 1249.996620 | probeB | 3a | 29999.918880 | True | 202 | 368 | [TH, Eth, APN, POL, LP, DG, CA1, VISpm, nan] |
| 729445652 | 719161530 | 1249.999897 | probeC | 3a | 29999.997521 | True | 207 | 373 | [APN, NOT, MB, DG, SUB, VISp, nan] |
| 729445654 | 719161530 | 1249.996707 | probeD | 3a | 29999.920963 | True | 93 | 358 | [grey, VL, CA3, CA2, CA1, VISl, nan] |
| 729445656 | 719161530 | 1249.999979 | probeE | 3a | 29999.999500 | True | 138 | 370 | [PO, VPM, TH, LP, LGd, CA3, DG, CA1, VISal, nan] |
Querying across channels#
… or across channels.
cache.get_channels().head()
| ecephys_probe_id | local_index | probe_horizontal_position | probe_vertical_position | anterior_posterior_ccf_coordinate | dorsal_ventral_ccf_coordinate | left_right_ccf_coordinate | ecephys_structure_id | ecephys_structure_acronym | ecephys_session_id | lfp_sampling_rate | phase | sampling_rate | has_lfp_data | unit_count | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||
| 849705558 | 792645504 | 1 | 11 | 20 | 8165 | 3314 | 6862 | 215.0 | APN | 779839471 | 1250.001479 | 3a | 30000.035489 | True | 0 |
| 849705560 | 792645504 | 2 | 59 | 40 | 8162 | 3307 | 6866 | 215.0 | APN | 779839471 | 1250.001479 | 3a | 30000.035489 | True | 0 |
| 849705562 | 792645504 | 3 | 27 | 40 | 8160 | 3301 | 6871 | 215.0 | APN | 779839471 | 1250.001479 | 3a | 30000.035489 | True | 0 |
| 849705564 | 792645504 | 4 | 43 | 60 | 8157 | 3295 | 6875 | 215.0 | APN | 779839471 | 1250.001479 | 3a | 30000.035489 | True | 0 |
| 849705566 | 792645504 | 5 | 11 | 60 | 8155 | 3288 | 6879 | 215.0 | APN | 779839471 | 1250.001479 | 3a | 30000.035489 | True | 0 |
Querying across units#
… as well as for sorted units.
units = cache.get_units()
units.head()
| waveform_PT_ratio | waveform_amplitude | amplitude_cutoff | cumulative_drift | d_prime | waveform_duration | ecephys_channel_id | firing_rate | waveform_halfwidth | isi_violations | isolation_distance | L_ratio | max_drift | nn_hit_rate | nn_miss_rate | presence_ratio | waveform_recovery_slope | waveform_repolarization_slope | silhouette_score | snr | waveform_spread | waveform_velocity_above | waveform_velocity_below | ecephys_probe_id | local_index | probe_horizontal_position | probe_vertical_position | anterior_posterior_ccf_coordinate | dorsal_ventral_ccf_coordinate | left_right_ccf_coordinate | ecephys_structure_id | ecephys_structure_acronym | ecephys_session_id | lfp_sampling_rate | name | phase | sampling_rate | has_lfp_data | date_of_acquisition | published_at | specimen_id | session_type | age_in_days | sex | genotype | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||||||||||||||||||||||||||||
| 915956282 | 0.611816 | 164.878740 | 0.072728 | 309.71 | 3.910873 | 0.535678 | 850229419 | 6.519432 | 0.164824 | 0.104910 | 30.546900 | 0.013865 | 27.10 | 0.898126 | 0.001599 | 0.99 | -0.087545 | 0.480915 | 0.102369 | 1.911839 | 30.0 | 0.000000 | NaN | 733744647 | 3 | 27 | 40 | -1000 | -1000 | -1000 | 8.0 | grey | 732592105 | 1249.996475 | probeB | 3a | 29999.915391 | True | 2019-01-09T00:26:20Z | 2019-10-03T00:00:00Z | 717038288 | brain_observatory_1.1 | 100.0 | M | wt/wt |
| 915956340 | 0.439372 | 247.254345 | 0.000881 | 160.24 | 5.519024 | 0.563149 | 850229419 | 9.660554 | 0.206030 | 0.006825 | 59.613182 | 0.000410 | 7.79 | 0.987654 | 0.000903 | 0.99 | -0.104196 | 0.704522 | 0.197458 | 3.357908 | 30.0 | 0.000000 | NaN | 733744647 | 3 | 27 | 40 | -1000 | -1000 | -1000 | 8.0 | grey | 732592105 | 1249.996475 | probeB | 3a | 29999.915391 | True | 2019-01-09T00:26:20Z | 2019-10-03T00:00:00Z | 717038288 | brain_observatory_1.1 | 100.0 | M | wt/wt |
| 915956345 | 0.500520 | 251.275830 | 0.001703 | 129.36 | 3.559911 | 0.521943 | 850229419 | 12.698430 | 0.192295 | 0.044936 | 47.805714 | 0.008281 | 11.56 | 0.930000 | 0.004956 | 0.99 | -0.153127 | 0.781296 | 0.138827 | 3.362198 | 30.0 | 0.343384 | NaN | 733744647 | 3 | 27 | 40 | -1000 | -1000 | -1000 | 8.0 | grey | 732592105 | 1249.996475 | probeB | 3a | 29999.915391 | True | 2019-01-09T00:26:20Z | 2019-10-03T00:00:00Z | 717038288 | brain_observatory_1.1 | 100.0 | M | wt/wt |
| 915956349 | 0.424620 | 177.115380 | 0.096378 | 169.29 | 2.973959 | 0.508208 | 850229419 | 16.192413 | 0.192295 | 0.120715 | 54.635515 | 0.010406 | 14.87 | 0.874667 | 0.021636 | 0.99 | -0.086022 | 0.553393 | 0.136901 | 2.684636 | 40.0 | 0.206030 | NaN | 733744647 | 3 | 27 | 40 | -1000 | -1000 | -1000 | 8.0 | grey | 732592105 | 1249.996475 | probeB | 3a | 29999.915391 | True | 2019-01-09T00:26:20Z | 2019-10-03T00:00:00Z | 717038288 | brain_observatory_1.1 | 100.0 | M | wt/wt |
| 915956356 | 0.512847 | 214.954545 | 0.054706 | 263.01 | 2.936851 | 0.549414 | 850229419 | 2.193113 | 0.233501 | 0.430427 | 18.136302 | 0.061345 | 18.37 | 0.637363 | 0.000673 | 0.99 | -0.106051 | 0.632977 | 0.108867 | 2.605408 | 60.0 | -0.451304 | NaN | 733744647 | 3 | 27 | 40 | -1000 | -1000 | -1000 | 8.0 | grey | 732592105 | 1249.996475 | probeB | 3a | 29999.915391 | True | 2019-01-09T00:26:20Z | 2019-10-03T00:00:00Z | 717038288 | brain_observatory_1.1 | 100.0 | M | wt/wt |
# There are quite a few of these
print(units.shape[0])
40010
Surveying metadata#
You can answer questions like: “what mouse genotypes were used in this dataset?” using your EcephysProjectCache.
print(f"stimulus sets: {cache.get_all_session_types()}")
print(f"genotypes: {cache.get_all_full_genotypes()}")
print(f"structures: {cache.get_structure_acronyms()}")
stimulus sets: ['brain_observatory_1.1', 'functional_connectivity']
genotypes: ['Sst-IRES-Cre/wt;Ai32(RCL-ChR2(H134R)_EYFP)/wt', 'Pvalb-IRES-Cre/wt;Ai32(RCL-ChR2(H134R)_EYFP)/wt', 'wt/wt', 'Vip-IRES-Cre/wt;Ai32(RCL-ChR2(H134R)_EYFP)/wt']
structures: ['APN', 'LP', 'MB', 'DG', 'CA1', 'VISrl', nan, 'TH', 'LGd', 'CA3', 'VIS', 'CA2', 'ProS', 'VISp', 'POL', 'VISpm', 'PPT', 'OP', 'NOT', 'HPF', 'SUB', 'VISam', 'ZI', 'LGv', 'VISal', 'VISl', 'SGN', 'SCig', 'MGm', 'MGv', 'VPM', 'grey', 'Eth', 'VPL', 'IGL', 'PP', 'PIL', 'PO', 'VISmma', 'POST', 'SCop', 'SCsg', 'SCzo', 'SCiw', 'IntG', 'MGd', 'MRN', 'LD', 'VISmmp', 'CP', 'VISli', 'PRE', 'RPF', 'LT', 'PF', 'PoT', 'VL', 'RT']
In order to look up a brain structure acronym, you can use our online atlas viewer. The AllenSDK additionally supports programmatic access to structure annotations. For more information, see the reference space and mouse connectivity documentation.
Obtaining an EcephysSession#
We package each session’s data into a Neurodata Without Borders 2.0 (NWB) file.
Calling get_session_data on your EcephysProjectCache will download such a
file and return an EcephysSession object.
EcephysSession objects contain methods and properties that access the data
within an ecephys NWB file and cache it in memory.
session_id = 756029989 # for example
session = cache.get_session_data(session_id)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'hdmf-common' version 1.1.3 because version 1.8.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.2.2 because version 2.7.0 is already loaded.
return func(args[0], **pargs)
This session object has some important metadata, such as the date and time at which the recording session started:
print(f"session {session.ecephys_session_id} was acquired in {session.session_start_time}")
session 756029989 was acquired in 2018-10-26 12:59:18-07:00
We’ll now jump in to accessing our session’s data. If you ever want a complete
documented list of the attributes and methods defined on EcephysSession, you
can run help(EcephysSession) (or in a jupyter notebook: EcephysSession?).
Sorted units#
Units are putative neurons, clustered from raw voltage traces using Kilosort 2. Each unit is associated with a single peak channel on a single probe, though its spikes might be picked up with some attenuation on multiple nearby channels. Each unit is assigned a unique integer identifier (“unit_id”) which can be used to look up its spike times and its mean waveform.
The units for a session are recorded in an attribute called, fittingly, units.
This is a pandas.DataFrame whose index is the unit id and whose columns
contain summary information about the unit, its peak channel, and its associated
probe.
session.units.head()
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'hdmf-common' version 1.1.3 because version 1.8.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.2.2 because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'hdmf-common' version 1.1.3 because version 1.8.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.2.2 because version 2.7.0 is already loaded.
return func(args[0], **pargs)
| waveform_PT_ratio | waveform_amplitude | amplitude_cutoff | cluster_id | cumulative_drift | d_prime | firing_rate | isi_violations | isolation_distance | L_ratio | local_index | max_drift | nn_hit_rate | nn_miss_rate | peak_channel_id | presence_ratio | waveform_recovery_slope | waveform_repolarization_slope | silhouette_score | snr | waveform_spread | waveform_velocity_above | waveform_velocity_below | waveform_duration | filtering | probe_channel_number | probe_horizontal_position | probe_id | probe_vertical_position | structure_acronym | ecephys_structure_id | ecephys_structure_acronym | anterior_posterior_ccf_coordinate | dorsal_ventral_ccf_coordinate | left_right_ccf_coordinate | probe_description | location | probe_sampling_rate | probe_lfp_sampling_rate | probe_has_lfp_data | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| unit_id | ||||||||||||||||||||||||||||||||||||||||
| 951814884 | 0.522713 | 187.434780 | 0.042404 | 6 | 463.86 | 3.555518 | 9.492176 | 0.181638 | 46.750473 | 0.024771 | 5 | 45.64 | 0.727333 | 0.016202 | 850126382 | 0.99 | -0.249885 | 0.673650 | 0.033776 | 3.342535 | 40.0 | 0.000000 | 0.000000 | 0.151089 | AP band: 500 Hz high-pass; LFP band: 1000 Hz l... | 4 | 43 | 760640083 | 60 | APN | 215.0 | APN | 8162 | 3487 | 6737 | probeA | See electrode locations | 29999.949611 | 1249.9979 | True |
| 951814876 | 0.652514 | 129.686505 | 0.097286 | 5 | 325.21 | 4.445414 | 39.100557 | 0.004799 | 85.178750 | 0.001785 | 4 | 40.68 | 1.000000 | 0.003756 | 850126382 | 0.99 | -0.143762 | 0.518633 | 0.108908 | 2.589717 | 50.0 | 0.000000 | 0.000000 | 0.315913 | AP band: 500 Hz high-pass; LFP band: 1000 Hz l... | 4 | 43 | 760640083 | 60 | APN | 215.0 | APN | 8162 | 3487 | 6737 | probeA | See electrode locations | 29999.949611 | 1249.9979 | True |
| 951815032 | 0.484297 | 207.380940 | 0.015482 | 17 | 396.28 | 3.848256 | 28.383277 | 0.007099 | 89.608836 | 0.035654 | 15 | 40.01 | 0.986000 | 0.014673 | 850126398 | 0.99 | -0.255492 | 0.766347 | 0.096715 | 3.811566 | 80.0 | -0.206030 | -0.686767 | 0.164824 | AP band: 500 Hz high-pass; LFP band: 1000 Hz l... | 12 | 43 | 760640083 | 140 | APN | 215.0 | APN | 8129 | 3419 | 6762 | probeA | See electrode locations | 29999.949611 | 1249.9979 | True |
| 951815275 | 0.600600 | 158.158650 | 0.063807 | 30 | 374.82 | 3.065938 | 5.709358 | 0.032317 | 48.114336 | 0.016783 | 27 | 33.32 | 0.883598 | 0.003683 | 850126416 | 0.99 | -0.206676 | 0.628944 | 0.144249 | 2.918134 | 60.0 | 0.686767 | 0.000000 | 0.178559 | AP band: 500 Hz high-pass; LFP band: 1000 Hz l... | 21 | 11 | 760640083 | 220 | APN | 215.0 | APN | 8095 | 3349 | 6787 | probeA | See electrode locations | 29999.949611 | 1249.9979 | True |
| 951815314 | 0.459025 | 173.475705 | 0.072129 | 34 | 420.05 | 4.198612 | 23.902235 | 0.048075 | 76.916334 | 0.009666 | 31 | 42.80 | 0.968000 | 0.017600 | 850126420 | 0.99 | -0.171503 | 0.740222 | 0.111106 | 3.360324 | 90.0 | -0.068677 | -0.274707 | 0.178559 | AP band: 500 Hz high-pass; LFP band: 1000 Hz l... | 23 | 27 | 760640083 | 240 | APN | 215.0 | APN | 8088 | 3333 | 6792 | probeA | See electrode locations | 29999.949611 | 1249.9979 | True |
As a pandas.DataFrame the units table supports many straightforward filtering
operations:
# how many units have signal to noise ratios that are greater than 4?
print(f'{session.units.shape[0]} units total')
units_with_very_high_snr = session.units[session.units['snr'] > 4]
print(f'{units_with_very_high_snr.shape[0]} units have snr > 4')
684 units total
81 units have snr > 4
… as well as some more advanced (and very useful!) operations. For more information, please see the pandas documentation. The following topics might be particularly handy:
Stimulus presentations#
During the course of a session, visual stimuli are presented on a monitor to the subject. We call intervals of time where a specific stimulus is presented (and its parameters held constant!) a stimulus presentation.
You can find information about the stimulus presentations that were displayed during a session by accessing the stimulus_presentations attribute on your EcephysSession object.
session.stimulus_presentations.head()
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'hdmf-common' version 1.1.3 because version 1.8.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.2.2 because version 2.7.0 is already loaded.
return func(args[0], **pargs)
| stimulus_block | start_time | stop_time | y_position | spatial_frequency | phase | stimulus_name | color | temporal_frequency | contrast | x_position | size | orientation | frame | duration | stimulus_condition_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| stimulus_presentation_id | ||||||||||||||||
| 0 | null | 24.429348 | 84.496188 | null | null | null | spontaneous | null | null | null | null | null | null | null | 60.066840 | 0 |
| 1 | 0.0 | 84.496188 | 84.729704 | 30.0 | 0.08 | [3644.93333333, 3644.93333333] | gabors | null | 4.0 | 0.8 | 40.0 | [20.0, 20.0] | 45.0 | null | 0.233516 | 1 |
| 2 | 0.0 | 84.729704 | 84.979900 | 10.0 | 0.08 | [3644.93333333, 3644.93333333] | gabors | null | 4.0 | 0.8 | -30.0 | [20.0, 20.0] | 45.0 | null | 0.250196 | 2 |
| 3 | 0.0 | 84.979900 | 85.230095 | -10.0 | 0.08 | [3644.93333333, 3644.93333333] | gabors | null | 4.0 | 0.8 | 10.0 | [20.0, 20.0] | 90.0 | null | 0.250196 | 3 |
| 4 | 0.0 | 85.230095 | 85.480291 | 40.0 | 0.08 | [3644.93333333, 3644.93333333] | gabors | null | 4.0 | 0.8 | 30.0 | [20.0, 20.0] | 90.0 | null | 0.250196 | 4 |
Like the units table, this is a pandas.DataFrame. Each row corresponds to a stimulus presentation and lists the time (on the session’s master clock, in seconds) when that presentation began and ended as well as the kind of stimulus that was presented (the “stimulus_name” column) and the parameter values that were used for that presentation. Many of these parameter values don’t overlap between stimulus classes, so the stimulus_presentations table uses the string "null" to indicate an inapplicable parameter. The index is named “stimulus_presentation_id” and many methods on EcephysSession use these ids.
Some of the columns bear a bit of explanation:
stimulus_block : A block consists of multiple presentations of the same stimulus class presented with (probably) different parameter values. If during a session stimuli were presented in the following order:
drifting gratings
static gratings
drifting gratings then the blocks for that session would be [0, 1, 2]. The gray period stimulus (just a blank gray screen) never gets a block.
duration : this is just stop_time - start_time, precalculated for convenience.
What kinds of stimuli were presented during this session? Pandas makes it easy to find out:
session.stimulus_names # just the unique values from the 'stimulus_name' column
['spontaneous',
'gabors',
'flashes',
'drifting_gratings',
'natural_movie_three',
'natural_movie_one',
'static_gratings',
'natural_scenes']
We can also obtain the stimulus epochs - blocks of time for which a particular
kind of stimulus was presented - for this session.
session.get_stimulus_epochs()
| start_time | stop_time | duration | stimulus_name | stimulus_block | |
|---|---|---|---|---|---|
| 0 | 24.429348 | 84.496188 | 60.066840 | spontaneous | null |
| 1 | 84.496188 | 996.491813 | 911.995625 | gabors | 0.0 |
| 2 | 996.491813 | 1285.483398 | 288.991585 | spontaneous | null |
| 3 | 1285.483398 | 1583.982946 | 298.499548 | flashes | 1.0 |
| 4 | 1583.982946 | 1585.734418 | 1.751472 | spontaneous | null |
| 5 | 1585.734418 | 2185.235561 | 599.501143 | drifting_gratings | 2.0 |
| 6 | 2185.235561 | 2216.261498 | 31.025937 | spontaneous | null |
| 7 | 2216.261498 | 2816.763498 | 600.502000 | natural_movie_three | 3.0 |
| 8 | 2816.763498 | 2846.788598 | 30.025100 | spontaneous | null |
| 9 | 2846.788598 | 3147.039578 | 300.250980 | natural_movie_one | 4.0 |
| 10 | 3147.039578 | 3177.064688 | 30.025110 | spontaneous | null |
| 11 | 3177.064688 | 3776.565851 | 599.501163 | drifting_gratings | 5.0 |
| 12 | 3776.565851 | 4077.834348 | 301.268497 | spontaneous | null |
| 13 | 4077.834348 | 4678.336348 | 600.502000 | natural_movie_three | 6.0 |
| 14 | 4678.336348 | 4708.361438 | 30.025090 | spontaneous | null |
| 15 | 4708.361438 | 5397.937871 | 689.576433 | drifting_gratings | 7.0 |
| 16 | 5397.937871 | 5398.938718 | 1.000847 | spontaneous | null |
| 17 | 5398.938718 | 5879.340268 | 480.401550 | static_gratings | 8.0 |
| 18 | 5879.340268 | 5909.365398 | 30.025130 | spontaneous | null |
| 19 | 5909.365398 | 6389.766968 | 480.401570 | natural_scenes | 9.0 |
| 20 | 6389.766968 | 6690.017948 | 300.250980 | spontaneous | null |
| 21 | 6690.017948 | 7170.419568 | 480.401620 | natural_scenes | 10.0 |
| 22 | 7170.419568 | 7200.444628 | 30.025060 | spontaneous | null |
| 23 | 7200.444628 | 7680.846188 | 480.401560 | static_gratings | 11.0 |
| 24 | 7680.846188 | 7710.871348 | 30.025160 | spontaneous | null |
| 25 | 7710.871348 | 8011.122288 | 300.250940 | natural_movie_one | 12.0 |
| 26 | 8011.122288 | 8041.147408 | 30.025120 | spontaneous | null |
| 27 | 8041.147408 | 8569.088694 | 527.941286 | natural_scenes | 13.0 |
| 28 | 8569.088694 | 8611.624248 | 42.535554 | spontaneous | null |
| 29 | 8611.624248 | 9152.076028 | 540.451780 | static_gratings | 14.0 |
If you are only interested in a subset of stimuli, you can either filter using
pandas or using the get_stimulus_table convience method:
session.get_stimulus_table(['drifting_gratings']).head()
| stimulus_block | start_time | stop_time | spatial_frequency | phase | stimulus_name | temporal_frequency | contrast | size | orientation | duration | stimulus_condition_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| stimulus_presentation_id | ||||||||||||
| 3798 | 2.0 | 1585.734418 | 1587.736098 | 0.04 | [42471.86666667, 42471.86666667] | drifting_gratings | 2.0 | 0.8 | [250.0, 250.0] | 180.0 | 2.00168 | 246 |
| 3799 | 2.0 | 1588.736891 | 1590.738571 | 0.04 | [42471.86666667, 42471.86666667] | drifting_gratings | 2.0 | 0.8 | [250.0, 250.0] | 135.0 | 2.00168 | 247 |
| 3800 | 2.0 | 1591.739398 | 1593.741078 | 0.04 | [42471.86666667, 42471.86666667] | drifting_gratings | 2.0 | 0.8 | [250.0, 250.0] | 180.0 | 2.00168 | 246 |
| 3801 | 2.0 | 1594.741921 | 1596.743591 | 0.04 | [42471.86666667, 42471.86666667] | drifting_gratings | 2.0 | 0.8 | [250.0, 250.0] | 270.0 | 2.00167 | 248 |
| 3802 | 2.0 | 1597.744458 | 1599.746088 | 0.04 | [42471.86666667, 42471.86666667] | drifting_gratings | 4.0 | 0.8 | [250.0, 250.0] | 135.0 | 2.00163 | 249 |
We might also want to know what the total set of available parameters is. The
get_stimulus_parameter_values method provides a dictionary mapping stimulus
parameters to the set of values that were applied to those parameters:
for key, values in session.get_stimulus_parameter_values().items():
print(f'{key}: {values}')
y_position: [30.0 10.0 -10.0 40.0 -40.0 -20.0 0.0 20.0 -30.0]
spatial_frequency: ['0.08' '[0.0, 0.0]' '0.04' 0.08 0.04 0.32 0.02 0.16]
phase: ['[3644.93333333, 3644.93333333]' '[0.0, 0.0]'
'[42471.86666667, 42471.86666667]' '0.0' '0.25' '0.5' '0.75']
color: [-1.0 1.0]
temporal_frequency: [4.0 2.0 15.0 1.0 8.0]
contrast: [0.8 1.0]
x_position: [40.0 -30.0 10.0 30.0 0.0 -40.0 -20.0 -10.0 20.0]
size: ['[20.0, 20.0]' '[300.0, 300.0]' '[250.0, 250.0]' '[1920.0, 1080.0]']
orientation: [45.0 90.0 0.0 180.0 135.0 270.0 315.0 225.0 30.0 150.0 120.0 60.0]
frame: [0.0 1.0 2.0 ... 3598.0 3599.0 -1.0]
Each distinct state of the monitor is called a “stimulus condition”. Each presentation in the stimulus presentations table exemplifies such a condition. This is encoded in its stimulus_condition_id field.
To get the full list of conditions presented in a session, use the stimulus_conditions attribute:
session.stimulus_conditions.head()
| y_position | spatial_frequency | mask | phase | stimulus_name | color | units | temporal_frequency | contrast | x_position | size | opacity | orientation | frame | color_triplet | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| stimulus_condition_id | |||||||||||||||
| 0 | null | null | null | null | spontaneous | null | null | null | null | null | null | null | null | null | null |
| 1 | 30.0 | 0.08 | circle | [3644.93333333, 3644.93333333] | gabors | null | deg | 4.0 | 0.8 | 40.0 | [20.0, 20.0] | True | 45.0 | null | [1.0, 1.0, 1.0] |
| 2 | 10.0 | 0.08 | circle | [3644.93333333, 3644.93333333] | gabors | null | deg | 4.0 | 0.8 | -30.0 | [20.0, 20.0] | True | 45.0 | null | [1.0, 1.0, 1.0] |
| 3 | -10.0 | 0.08 | circle | [3644.93333333, 3644.93333333] | gabors | null | deg | 4.0 | 0.8 | 10.0 | [20.0, 20.0] | True | 90.0 | null | [1.0, 1.0, 1.0] |
| 4 | 40.0 | 0.08 | circle | [3644.93333333, 3644.93333333] | gabors | null | deg | 4.0 | 0.8 | 30.0 | [20.0, 20.0] | True | 90.0 | null | [1.0, 1.0, 1.0] |
Spike data#
The EcephysSession object holds spike times (in seconds on the session master
clock) for each unit. These are stored in a dictionary, which maps unit ids (the
index values of the units table) to arrays of spike times.
# grab an arbitrary (though high-snr!) unit (we made units_with_high_snr above)
high_snr_unit_ids = units_with_very_high_snr.index.values
unit_id = high_snr_unit_ids[0]
print(f"{len(session.spike_times[unit_id])} spikes were detected for unit {unit_id} at times:")
session.spike_times[unit_id]
236169 spikes were detected for unit 951816951 at times:
array([3.81328401e+00, 4.20301799e+00, 4.30151816e+00, ...,
9.96615988e+03, 9.96617945e+03, 9.96619655e+03])
You can also obtain spikes tagged with the stimulus presentation during which they occurred:
# get spike times from the first block of drifting gratings presentations
drifting_gratings_presentation_ids = session.stimulus_presentations.loc[
(session.stimulus_presentations['stimulus_name'] == 'drifting_gratings')
].index.values
times = session.presentationwise_spike_times(
stimulus_presentation_ids=drifting_gratings_presentation_ids,
unit_ids=high_snr_unit_ids
)
times.head()
| stimulus_presentation_id | unit_id | time_since_stimulus_presentation_onset | |
|---|---|---|---|
| spike_time | |||
| 1585.734841 | 3798 | 951817927 | 0.000423 |
| 1585.736862 | 3798 | 951812742 | 0.002444 |
| 1585.738591 | 3798 | 951805427 | 0.004173 |
| 1585.738941 | 3798 | 951816951 | 0.004523 |
| 1585.738995 | 3798 | 951820510 | 0.004577 |
We can make raster plots of these data:
first_drifting_grating_presentation_id = times['stimulus_presentation_id'].values[0]
plot_times = times[times['stimulus_presentation_id'] == first_drifting_grating_presentation_id]
fig = raster_plot(plot_times, title=f'spike raster for stimulus presentation {first_drifting_grating_presentation_id}')
plt.show()
# also print out this presentation
session.stimulus_presentations.loc[first_drifting_grating_presentation_id]
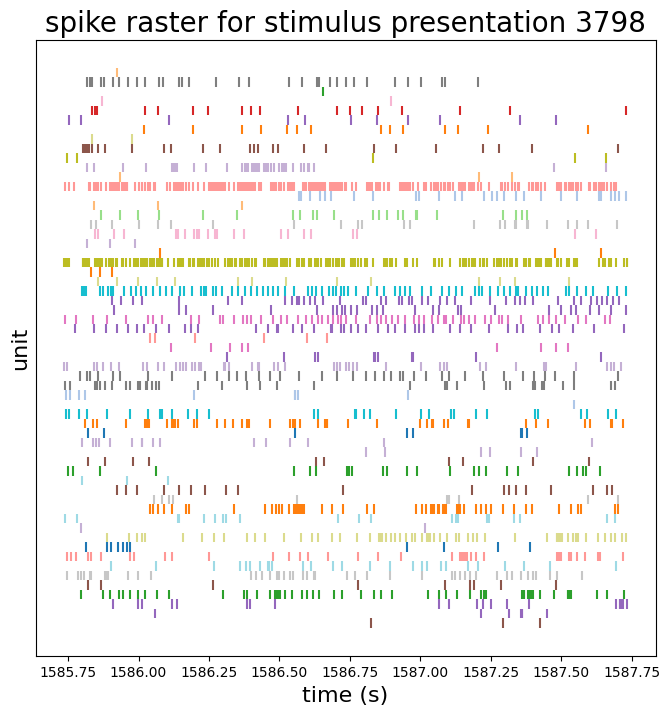
stimulus_block 2.0
start_time 1585.734418
stop_time 1587.736098
y_position null
spatial_frequency 0.04
phase [42471.86666667, 42471.86666667]
stimulus_name drifting_gratings
color null
temporal_frequency 2.0
contrast 0.8
x_position null
size [250.0, 250.0]
orientation 180.0
frame null
duration 2.00168
stimulus_condition_id 246
Name: 3798, dtype: object
We can access summary spike statistics for stimulus conditions and unit
stats = session.conditionwise_spike_statistics(
stimulus_presentation_ids=drifting_gratings_presentation_ids,
unit_ids=high_snr_unit_ids
)
# display the parameters associated with each condition
stats = pd.merge(stats, session.stimulus_conditions, left_on="stimulus_condition_id", right_index=True)
stats.head()
| spike_count | stimulus_presentation_count | spike_mean | spike_std | spike_sem | y_position | spatial_frequency | mask | phase | stimulus_name | color | units | temporal_frequency | contrast | x_position | size | opacity | orientation | frame | color_triplet | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| unit_id | stimulus_condition_id | ||||||||||||||||||||
| 951799336 | 246 | 13 | 15 | 0.866667 | 1.995232 | 0.515167 | null | 0.04 | None | [42471.86666667, 42471.86666667] | drifting_gratings | null | deg | 2.0 | 0.8 | null | [250.0, 250.0] | True | 180.0 | null | [1.0, 1.0, 1.0] |
| 951800977 | 246 | 26 | 15 | 1.733333 | 2.737743 | 0.706882 | null | 0.04 | None | [42471.86666667, 42471.86666667] | drifting_gratings | null | deg | 2.0 | 0.8 | null | [250.0, 250.0] | True | 180.0 | null | [1.0, 1.0, 1.0] |
| 951801127 | 246 | 103 | 15 | 6.866667 | 7.414914 | 1.914523 | null | 0.04 | None | [42471.86666667, 42471.86666667] | drifting_gratings | null | deg | 2.0 | 0.8 | null | [250.0, 250.0] | True | 180.0 | null | [1.0, 1.0, 1.0] |
| 951801187 | 246 | 4 | 15 | 0.266667 | 0.593617 | 0.153271 | null | 0.04 | None | [42471.86666667, 42471.86666667] | drifting_gratings | null | deg | 2.0 | 0.8 | null | [250.0, 250.0] | True | 180.0 | null | [1.0, 1.0, 1.0] |
| 951801462 | 246 | 83 | 15 | 5.533333 | 2.587516 | 0.668094 | null | 0.04 | None | [42471.86666667, 42471.86666667] | drifting_gratings | null | deg | 2.0 | 0.8 | null | [250.0, 250.0] | True | 180.0 | null | [1.0, 1.0, 1.0] |
Using these data, we can ask for each unit: which stimulus condition evoked the most activity on average?
with_repeats = stats[stats["stimulus_presentation_count"] >= 5]
highest_mean_rate = lambda df: df.loc[df['spike_mean'].idxmax()]
max_rate_conditions = with_repeats.groupby('unit_id').apply(highest_mean_rate)
max_rate_conditions.head()
| spike_count | stimulus_presentation_count | spike_mean | spike_std | spike_sem | y_position | spatial_frequency | mask | phase | stimulus_name | color | units | temporal_frequency | contrast | x_position | size | opacity | orientation | frame | color_triplet | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| unit_id | ||||||||||||||||||||
| 951799336 | 81 | 15 | 5.400000 | 9.287472 | 2.398015 | null | 0.04 | None | [42471.86666667, 42471.86666667] | drifting_gratings | null | deg | 4.0 | 0.8 | null | [250.0, 250.0] | True | 0.0 | null | [1.0, 1.0, 1.0] |
| 951800977 | 41 | 15 | 2.733333 | 2.840188 | 0.733333 | null | 0.04 | None | [42471.86666667, 42471.86666667] | drifting_gratings | null | deg | 1.0 | 0.8 | null | [250.0, 250.0] | True | 45.0 | null | [1.0, 1.0, 1.0] |
| 951801127 | 209 | 15 | 13.933333 | 9.728211 | 2.511813 | null | 0.04 | None | [42471.86666667, 42471.86666667] | drifting_gratings | null | deg | 1.0 | 0.8 | null | [250.0, 250.0] | True | 90.0 | null | [1.0, 1.0, 1.0] |
| 951801187 | 53 | 15 | 3.533333 | 5.902380 | 1.523988 | null | 0.04 | None | [42471.86666667, 42471.86666667] | drifting_gratings | null | deg | 4.0 | 0.8 | null | [250.0, 250.0] | True | 270.0 | null | [1.0, 1.0, 1.0] |
| 951801462 | 136 | 15 | 9.066667 | 5.161211 | 1.332619 | null | 0.04 | None | [42471.86666667, 42471.86666667] | drifting_gratings | null | deg | 2.0 | 0.8 | null | [250.0, 250.0] | True | 45.0 | null | [1.0, 1.0, 1.0] |
Spike histograms#
It is commonly useful to compare spike data from across units and stimulus
presentations, all relative to the onset of a stimulus presentation. We can do
this using the presentationwise_spike_counts method.
# We're going to build an array of spike counts surrounding stimulus presentation onset
# To do that, we will need to specify some bins (in seconds, relative to stimulus onset)
time_bin_edges = np.linspace(-0.01, 0.4, 200)
# look at responses to the flash stimulus
flash_250_ms_stimulus_presentation_ids = session.stimulus_presentations[
session.stimulus_presentations['stimulus_name'] == 'flashes'
].index.values
# and get a set of units with only decent snr
decent_snr_unit_ids = session.units[
session.units['snr'] >= 1.5
].index.values
spike_counts_da = session.presentationwise_spike_counts(
bin_edges=time_bin_edges,
stimulus_presentation_ids=flash_250_ms_stimulus_presentation_ids,
unit_ids=decent_snr_unit_ids
)
spike_counts_da
<xarray.DataArray 'spike_counts' (stimulus_presentation_id: 150,
time_relative_to_stimulus_onset: 199,
unit_id: 631)>
array([[[0, 1, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0],
...,
[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 1, 0, 0],
[0, 0, 0, ..., 0, 0, 0]],
[[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0],
...,
[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0]],
[[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0],
...,
...
...,
[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0]],
[[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0],
...,
[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0],
[1, 0, 0, ..., 0, 0, 0]],
[[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0],
...,
[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0]]], dtype=uint16)
Coordinates:
* stimulus_presentation_id (stimulus_presentation_id) int64 3647 .....
* time_relative_to_stimulus_onset (time_relative_to_stimulus_onset) float64 ...
* unit_id (unit_id) int64 951814884 ... 951814312This has returned a new (to this notebook) data structure, the
xarray.DataArray. You can think of this as similar to a 3+D
pandas.DataFrame, or as a numpy.ndarray with labeled axes and indices. See
the xarray documentation for
more information. In the mean time, the salient features are:
Dimensions : Each axis on each data variable is associated with a named dimension. This lets us see unambiguously what the axes of our array mean.
Coordinates : Arrays of labels for each sample on each dimension.
xarray is nice because it forces code to be explicit about dimensions and coordinates, improving readability and avoiding bugs. However, you can always convert to numpy or pandas data structures as follows:
to pandas:
spike_counts_ds.to_dataframe()produces a multiindexed dataframeto numpy:
spike_counts_ds.valuesgives you access to the underlying numpy array
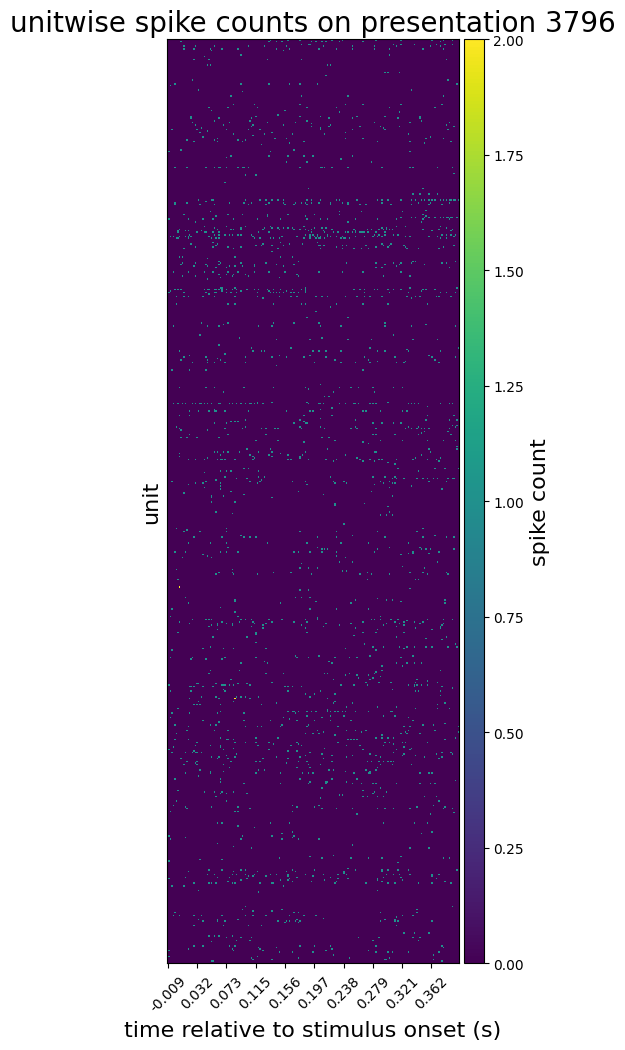
We can now plot spike counts for a particular presentation:
presentation_id = 3796 # chosen arbitrarily
plot_spike_counts(
spike_counts_da.loc[{'stimulus_presentation_id': presentation_id}],
spike_counts_da['time_relative_to_stimulus_onset'],
'spike count',
f'unitwise spike counts on presentation {presentation_id}'
)
plt.show()
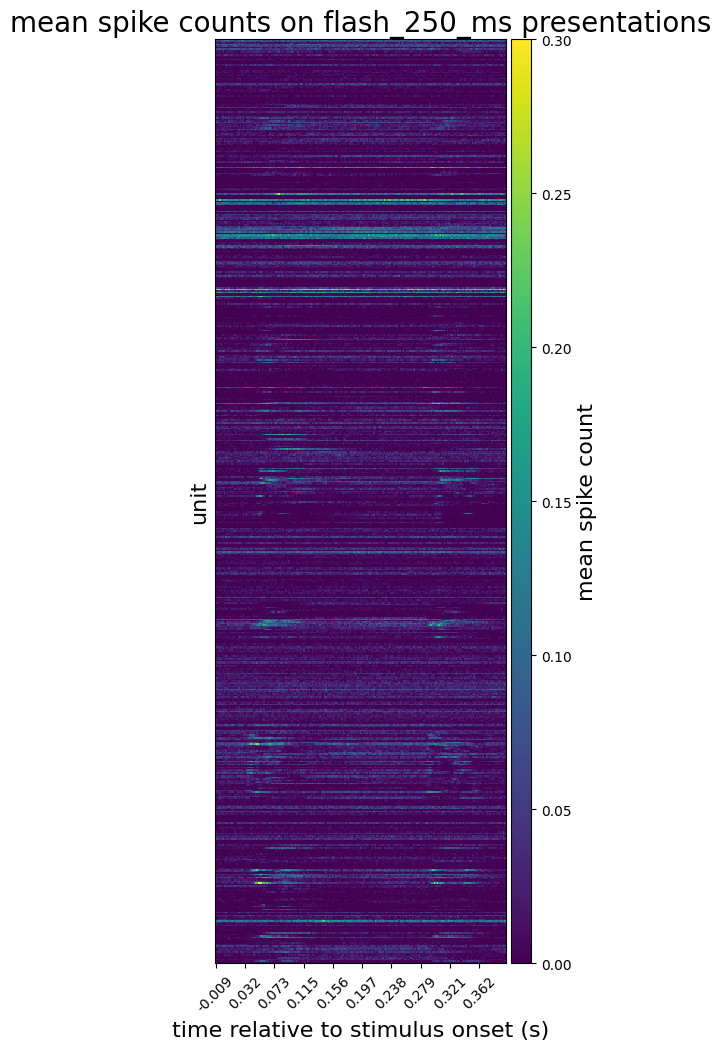
We can also average across all presentations, adding a new data array to the dataset. Notice that this one no longer has a stimulus_presentation_id dimension, as we have collapsed it by averaging.
mean_spike_counts = spike_counts_da.mean(dim='stimulus_presentation_id')
mean_spike_counts
<xarray.DataArray 'spike_counts' (time_relative_to_stimulus_onset: 199,
unit_id: 631)>
array([[0.02666667, 0.1 , 0.08 , ..., 0.00666667, 0. ,
0.00666667],
[0.02666667, 0.06666667, 0.04666667, ..., 0.02 , 0. ,
0. ],
[0.02 , 0.11333333, 0.03333333, ..., 0.03333333, 0. ,
0. ],
...,
[0.02666667, 0.09333333, 0.05333333, ..., 0.00666667, 0. ,
0. ],
[0.02666667, 0.06666667, 0.02666667, ..., 0.00666667, 0.02 ,
0. ],
[0.02666667, 0.12 , 0.02 , ..., 0. , 0. ,
0. ]])
Coordinates:
* time_relative_to_stimulus_onset (time_relative_to_stimulus_onset) float64 ...
* unit_id (unit_id) int64 951814884 ... 951814312… and plot the mean spike counts
plot_spike_counts(
mean_spike_counts,
mean_spike_counts['time_relative_to_stimulus_onset'],
'mean spike count',
'mean spike counts on flash_250_ms presentations'
)
plt.show()
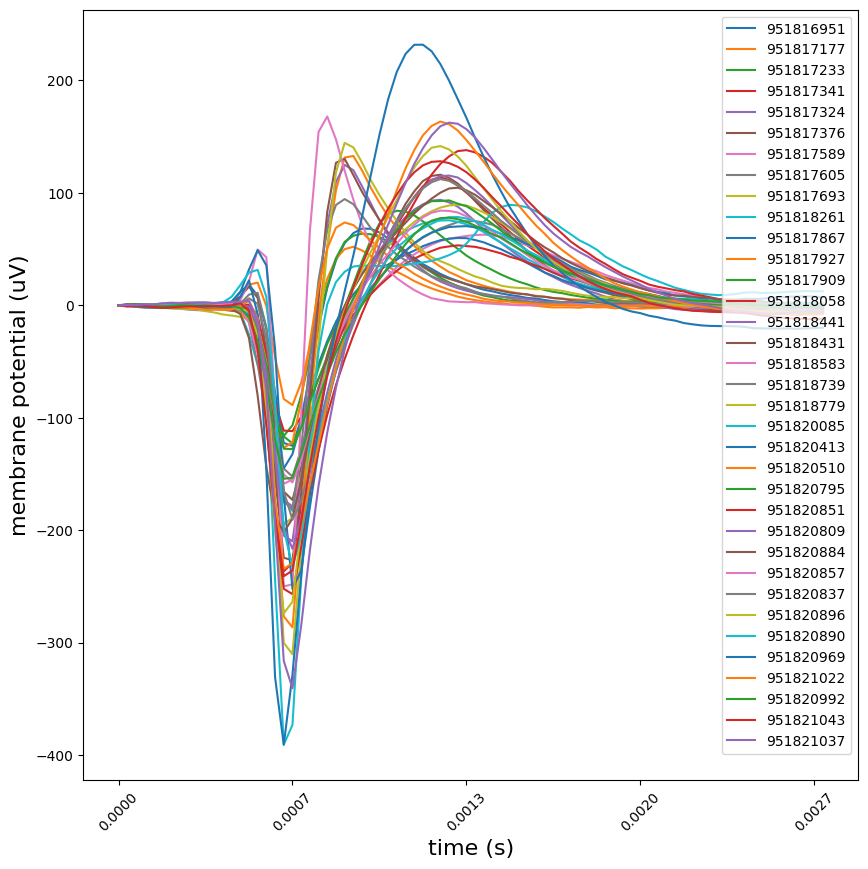
Waveforms#
We store precomputed mean waveforms for each unit in the mean_waveforms
attribute on the EcephysSession object. This is a dictionary which maps unit
ids to xarray
DataArrays.
These have channel and time (seconds, aligned to the detected event times)
dimensions. The data values are in microvolts, as measured at the recording
site.
units_of_interest = high_snr_unit_ids[:35]
waveforms = {uid: session.mean_waveforms[uid] for uid in units_of_interest}
peak_channels = {uid: session.units.loc[uid, 'peak_channel_id'] for uid in units_of_interest}
# plot the mean waveform on each unit's peak channel/
plot_mean_waveforms(waveforms, units_of_interest, peak_channels)
plt.show()
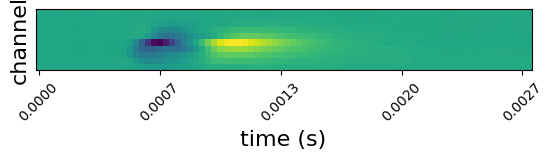
Since neuropixels probes are densely populated with channels, spikes are typically detected on several channels. We can see this by plotting mean waveforms on channels surrounding a unit’s peak channel:
uid = units_of_interest[12]
unit_waveforms = waveforms[uid]
peak_channel = peak_channels[uid]
peak_channel_idx = np.where(unit_waveforms["channel_id"] == peak_channel)[0][0]
ch_min = max(peak_channel_idx - 10, 0)
ch_max = min(peak_channel_idx + 10, len(unit_waveforms["channel_id"]) - 1)
surrounding_channels = unit_waveforms["channel_id"][np.arange(ch_min, ch_max, 2)]
fig, ax = plt.subplots()
ax.imshow(unit_waveforms.loc[{"channel_id": surrounding_channels}])
ax.yaxis.set_major_locator(plt.NullLocator())
ax.set_ylabel("channel", fontsize=16)
ax.set_xticks(np.arange(0, len(unit_waveforms['time']), 20))
ax.set_xticklabels([f'{float(ii):1.4f}' for ii in unit_waveforms['time'][::20]], rotation=45)
ax.set_xlabel("time (s)", fontsize=16)
plt.show()
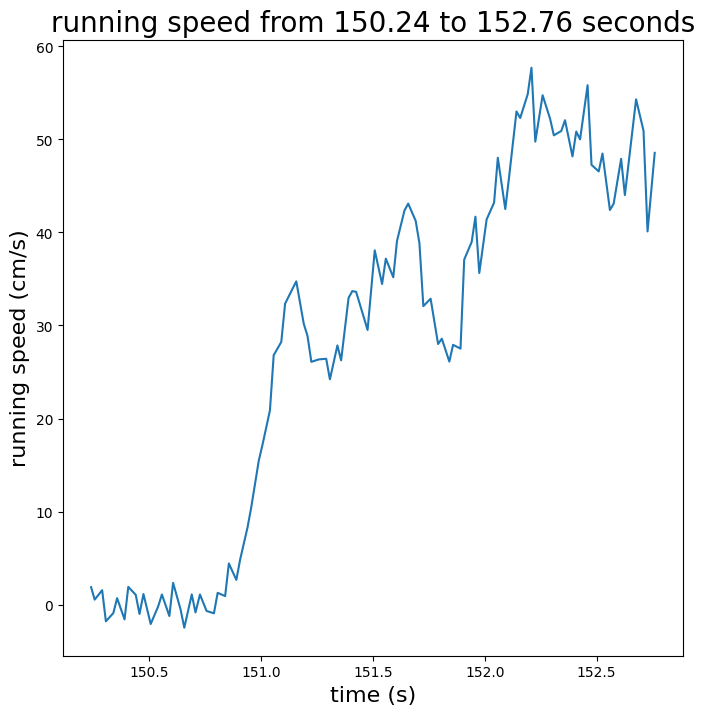
Running speed#
We can obtain the velocity at which the experimental subject ran as a function
of time by accessing the running_speed attribute. This returns a pandas
dataframe whose rows are intervals of time (defined by “start_time” and
“end_time” columns), and whose “velocity” column contains mean running speeds
within those intervals.
Here we’ll plot the running speed trace for an arbitrary chunk of time.
running_speed_midpoints = session.running_speed["start_time"] + \
(session.running_speed["end_time"] - session.running_speed["start_time"]) / 2
plot_running_speed(
running_speed_midpoints,
session.running_speed["velocity"],
start_index=5000,
stop_index=5100
)
plt.show()
Optogenetic stimulation#
session.optogenetic_stimulation_epochs
| start_time | condition | level | stop_time | stimulus_name | duration | |
|---|---|---|---|---|---|---|
| id | ||||||
| 0 | 9224.65339 | half-period of a cosine wave | 1.0 | 9225.65339 | raised_cosine | 1.000 |
| 1 | 9226.82361 | half-period of a cosine wave | 2.5 | 9227.82361 | raised_cosine | 1.000 |
| 2 | 9228.64353 | a single square pulse | 1.0 | 9228.64853 | pulse | 0.005 |
| 3 | 9230.65374 | a single square pulse | 4.0 | 9230.66374 | pulse | 0.010 |
| 4 | 9232.42375 | 2.5 ms pulses at 10 Hz | 4.0 | 9233.42375 | fast_pulses | 1.000 |
| ... | ... | ... | ... | ... | ... | ... |
| 175 | 9562.68831 | a single square pulse | 4.0 | 9562.69331 | pulse | 0.005 |
| 176 | 9564.75842 | a single square pulse | 1.0 | 9564.76842 | pulse | 0.010 |
| 177 | 9566.86855 | a single square pulse | 2.5 | 9566.87855 | pulse | 0.010 |
| 178 | 9568.98860 | a single square pulse | 4.0 | 9568.99860 | pulse | 0.010 |
| 179 | 9571.11868 | half-period of a cosine wave | 4.0 | 9572.11868 | raised_cosine | 1.000 |
180 rows × 6 columns
Eye tracking ellipse fits and estimated screen gaze location#
Ecephys sessions may contain eye tracking data in the form of ellipse fits and estimated screen gaze location. Let’s look at the ellipse fits first:
pupil_data = session.get_pupil_data()
pupil_data
| corneal_reflection_center_x | corneal_reflection_center_y | corneal_reflection_height | corneal_reflection_width | corneal_reflection_phi | pupil_center_x | pupil_center_y | pupil_height | pupil_width | pupil_phi | eye_center_x | eye_center_y | eye_height | eye_width | eye_phi | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time (s) | |||||||||||||||
| 3.20620 | 321.221602 | 215.484021 | 11.091658 | 12.801322 | -0.340513 | 291.258341 | 187.965473 | 96.294414 | 102.751684 | -0.759200 | 310.242901 | 207.161069 | 273.535714 | 323.227726 | 0.099357 |
| 3.22948 | 321.291022 | 215.282122 | 11.318211 | 12.807982 | -0.243113 | 290.703953 | 188.659872 | 94.239278 | 103.384550 | -0.715791 | 310.140402 | 207.007975 | 273.303505 | 322.919143 | 0.091675 |
| 3.23714 | 321.294868 | 215.055000 | 11.136122 | 12.423237 | -0.331511 | 290.687532 | 188.294754 | 94.729755 | 103.814759 | -0.673967 | 310.224041 | 206.700513 | 273.481027 | 323.497356 | 0.090202 |
| 3.27028 | 321.288914 | 215.250999 | 10.596050 | 12.414694 | -0.245863 | 289.154532 | 188.524943 | 93.877264 | 99.315115 | -0.732209 | 310.134502 | 206.791105 | 273.988038 | 323.147754 | 0.091154 |
| 3.30396 | 320.819438 | 215.915143 | 9.366927 | 12.870596 | 0.109020 | 290.206682 | 187.411271 | 90.221821 | 103.856061 | -0.583017 | 310.226498 | 206.519467 | 273.542559 | 322.233983 | 0.097546 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 9939.71240 | 322.580849 | 216.600349 | 12.799143 | 13.400555 | -0.525985 | NaN | NaN | NaN | NaN | NaN | 313.501794 | 210.587410 | 280.224381 | 322.533004 | 0.167348 |
| 9939.74576 | 324.007748 | 216.520870 | 11.843170 | 13.212379 | -0.403054 | NaN | NaN | NaN | NaN | NaN | 313.532165 | 211.023852 | 280.388767 | 322.942096 | 0.189335 |
| 9939.77918 | 324.118835 | 216.898871 | 11.336945 | 12.877454 | -0.254458 | NaN | NaN | NaN | NaN | NaN | 314.227284 | 210.967838 | 279.978946 | 323.751767 | 0.201428 |
| 9939.81257 | 324.141869 | 215.929826 | 12.343291 | 13.347376 | -0.186693 | NaN | NaN | NaN | NaN | NaN | 313.128124 | 210.027071 | 280.560120 | 321.832875 | 0.189717 |
| 9939.84647 | 324.200410 | 215.621956 | 13.629577 | 14.614380 | -0.270081 | NaN | NaN | NaN | NaN | NaN | 312.543325 | 209.802086 | 279.835031 | 321.055079 | 0.193363 |
297838 rows × 15 columns
This particular session has eye tracking data, let’s try plotting the ellipse fits over time.
%%capture
from matplotlib import animation
from matplotlib.patches import Ellipse
def plot_animated_ellipse_fits(pupil_data: pd.DataFrame, start_frame: int, end_frame: int):
start_frame = 0 if (start_frame < 0) else start_frame
end_frame = len(pupil_data) if (end_frame > len(pupil_data)) else end_frame
frame_times = pupil_data.index.values[start_frame:end_frame]
interval = np.average(np.diff(frame_times)) * 1000
fig = plt.figure()
ax = plt.axes(xlim=(0, 480), ylim=(0, 480))
cr_ellipse = Ellipse((0, 0), width=0.0, height=0.0, angle=0, color='white')
pupil_ellipse = Ellipse((0, 0), width=0.0, height=0.0, angle=0, color='black')
eye_ellipse = Ellipse((0, 0), width=0.0, height=0.0, angle=0, color='grey')
ax.add_patch(eye_ellipse)
ax.add_patch(pupil_ellipse)
ax.add_patch(cr_ellipse)
def update_ellipse(ellipse_patch, ellipse_frame_vals: pd.DataFrame, prefix: str):
ellipse_patch.center = tuple(ellipse_frame_vals[[f"{prefix}_center_x", f"{prefix}_center_y"]].values)
ellipse_patch.width = ellipse_frame_vals[f"{prefix}_width"]
ellipse_patch.height = ellipse_frame_vals[f"{prefix}_height"]
ellipse_patch.angle = np.degrees(ellipse_frame_vals[f"{prefix}_phi"])
def init():
return [cr_ellipse, pupil_ellipse, eye_ellipse]
def animate(i):
ellipse_frame_vals = pupil_data.iloc[i]
update_ellipse(cr_ellipse, ellipse_frame_vals, prefix="corneal_reflection")
update_ellipse(pupil_ellipse, ellipse_frame_vals, prefix="pupil")
update_ellipse(eye_ellipse, ellipse_frame_vals, prefix="eye")
return [cr_ellipse, pupil_ellipse, eye_ellipse]
return animation.FuncAnimation(fig, animate, init_func=init, interval=interval, frames=range(start_frame, end_frame), blit=True)
anim = plot_animated_ellipse_fits(pupil_data, 100, 600)
from IPython.display import HTML
HTML(anim.to_jshtml())
