Behavior Session Data#
This notebook shows how to access all behavior session data for one mouse and aggregate data across sessions to look at training history.
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib notebook
%matplotlib inline
# Import allenSDK and check the version, which should be 2.16.2
import allensdk
allensdk.__version__
'2.16.2'
# import the behavior ophys project cache class from SDK to be able to load the data
from allensdk.brain_observatory.behavior.behavior_project_cache import VisualBehaviorOphysProjectCache
/opt/envs/allensdk/lib/python3.10/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
Load the cache and get the behavior sessions table#
# Set the path to the dataset
cache_dir = '/root/capsule/data/'
# If you are working with data in the cloud in Code Ocean,
# or if you have already downloaded the full dataset to your local machine,
# you can instantiate a local cache
cache = VisualBehaviorOphysProjectCache.from_local_cache(cache_dir=cache_dir, use_static_cache=True)
# If you are working with the data locally for the first time, you need to instantiate the cache from S3:
# cache = VisualBehaviorOphysProjectCache.from_s3_cache(cache_dir=cache_dir)
behavior_session_table = cache.get_behavior_session_table()
/opt/envs/allensdk/lib/python3.10/site-packages/allensdk/brain_observatory/behavior/behavior_project_cache/behavior_project_cache.py:135: UpdatedStimulusPresentationTableWarning:
As of AllenSDK version 2.16.0, the latest Visual Behavior Ophys data has been significantly updated from previous releases. Specifically the user will need to update all processing of the stimulus_presentations tables. These tables now include multiple stimulus types delineated by the columns `stimulus_block` and `stimulus_block_name`.
The data that was available in previous releases are stored in the block name containing 'change_detection' and can be accessed in the pandas table by using:
`stimulus_presentations[stimulus_presentations.stimulus_block_name.str.contains('change_detection')]`
warnings.warn(
View a sample of the behavior session table#
The behavior_session_table is a Pandas DataFrame with one row for every behavior session and informative metadata columns.
The behavior_session_table includes sessions performed on a two-photon imaging rig (session_type starting with OPHYS_) and training sessions performed in the behavior facility (session_type starting with TRAINING_).
# view 10 randomly selected rows of the table using pandas sample command
behavior_session_table.sample(10)
| ophys_session_id | ophys_container_id | mouse_id | indicator | full_genotype | driver_line | cre_line | reporter_line | sex | age_in_days | ... | num_targeted_structures | catch_trial_count | engaged_trial_count | hit_trial_count | false_alarm_trial_count | correct_reject_trial_count | trial_count | go_trial_count | miss_trial_count | file_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| behavior_session_id | |||||||||||||||||||||
| 1103017152 | <NA> | NaN | 563234 | GCaMP6f | Vip-IRES-Cre/wt;Ai148(TIT2L-GC6f-ICL-tTA2)/wt | [Vip-IRES-Cre] | Vip-IRES-Cre | Ai148(TIT2L-GC6f-ICL-tTA2) | F | 164 | ... | <NA> | 33 | 490 | 129 | 6 | 27 | 626 | 228 | 99 | 4208 |
| 842399625 | <NA> | NaN | 436662 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | M | 125 | ... | <NA> | 33 | 493 | 133 | 9 | 24 | 719 | 231 | 98 | 4356 |
| 892413875 | <NA> | NaN | 462468 | GCaMP6s | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai94(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai94(TITL-GCaMP6s) | M | 92 | ... | <NA> | 34 | 518 | 104 | 4 | 30 | 693 | 238 | 134 | 4926 |
| 974606492 | 974479344 | [968451510] | 479426 | GCaMP6s | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai94(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai94(TITL-GCaMP6s) | F | 142 | ... | 1 | 27 | 619 | 164 | 1 | 26 | 649 | 197 | 33 | -999 |
| 951696355 | <NA> | NaN | 467953 | GCaMP6f | Vip-IRES-Cre/wt;Ai148(TIT2L-GC6f-ICL-tTA2)/wt | [Vip-IRES-Cre] | Vip-IRES-Cre | Ai148(TIT2L-GC6f-ICL-tTA2) | M | 157 | ... | <NA> | 19 | 204 | 80 | 3 | 16 | 805 | 147 | 67 | 2488 |
| 993570083 | 993550962 | [1022733783, 1022733786, 1022733792, 102273379... | 484631 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 155 | ... | 4 | 35 | 310 | 103 | 3 | 32 | 792 | 248 | 145 | -999 |
| 949348713 | 949217880 | [1022731524, 1022731527, 1022731531, 102273153... | 453991 | GCaMP6f | Vip-IRES-Cre/wt;Ai148(TIT2L-GC6f-ICL-tTA2)/wt | [Vip-IRES-Cre] | Vip-IRES-Cre | Ai148(TIT2L-GC6f-ICL-tTA2) | M | 222 | ... | 2 | 50 | 0 | 0 | 0 | 50 | 401 | 351 | 351 | -999 |
| 779111633 | <NA> | NaN | 421136 | GCaMP6f | Vip-IRES-Cre/wt;Ai148(TIT2L-GC6f-ICL-tTA2)/wt | [Vip-IRES-Cre] | Vip-IRES-Cre | Ai148(TIT2L-GC6f-ICL-tTA2) | M | 76 | ... | <NA> | 46 | 205 | 115 | 1 | 45 | 609 | 308 | 193 | 5112 |
| 998337326 | <NA> | NaN | 489065 | GCaMP6f | Vip-IRES-Cre/wt;Ai148(TIT2L-GC6f-ICL-tTA2)/wt | [Vip-IRES-Cre] | Vip-IRES-Cre | Ai148(TIT2L-GC6f-ICL-tTA2) | F | 156 | ... | <NA> | 20 | 226 | 82 | 8 | 12 | 905 | 125 | 43 | 5759 |
| 870481685 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 85 | ... | <NA> | 40 | 318 | 128 | 3 | 37 | 537 | 285 | 157 | 4775 |
10 rows × 34 columns
Select a mouse for analysis#
We’ll choose one mouse id from the full list of unique mouse IDs in the dataset
mouse_id = behavior_session_table['mouse_id'].unique()[76]
mouse_id
'456915'
Query the full behavior sessions table for all sessions that this mouse performed#
This will return a subset of the full behavior_session_table in which the mouse_id matches our mouse_id variable (mouse 445002). The table should be returned in order of date of acquisition, but we’ll use the Pandas command sort_values(by = 'date_of_acquisition') just to be sure.
What we then see is a table that has metadata for every session performed by this mouse, in sequential order. The equipment_name column tells us where the session was run on that day and the session_type column tells us the name of the session type. See the technical white paper for a detailed description of the progression of stages.
this_mouse_table = behavior_session_table.query('mouse_id == @mouse_id').sort_values(by = 'date_of_acquisition')
# note that the following is functionally equivalent if you find the syntax easier to read:
# this_mouse_table = behavior_session_table[behavior_session_table['mouse_id'] == mouse_id]
this_mouse_table
Show code cell output
| ophys_session_id | ophys_container_id | mouse_id | indicator | full_genotype | driver_line | cre_line | reporter_line | sex | age_in_days | ... | num_targeted_structures | catch_trial_count | engaged_trial_count | hit_trial_count | false_alarm_trial_count | correct_reject_trial_count | trial_count | go_trial_count | miss_trial_count | file_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| behavior_session_id | |||||||||||||||||||||
| 862245493 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 71 | ... | <NA> | 0 | 108 | 0 | 0 | 0 | 118 | 0 | 0 | 4765 |
| 863368489 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 74 | ... | <NA> | 26 | 254 | 55 | 6 | 20 | 1466 | 176 | 121 | 4766 |
| 863966974 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 75 | ... | <NA> | 29 | 352 | 78 | 5 | 24 | 1175 | 175 | 97 | 4767 |
| 864665204 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 76 | ... | <NA> | 39 | 232 | 86 | 7 | 32 | 786 | 246 | 160 | 4768 |
| 865131331 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 77 | ... | <NA> | 39 | 182 | 86 | 5 | 34 | 633 | 306 | 220 | 4769 |
| 865887821 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 78 | ... | <NA> | 39 | 265 | 91 | 3 | 36 | 595 | 256 | 165 | 4770 |
| 866909432 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 81 | ... | <NA> | 25 | 289 | 103 | 5 | 20 | 797 | 201 | 98 | 4771 |
| 867521015 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 82 | ... | <NA> | 34 | 355 | 101 | 1 | 33 | 688 | 242 | 141 | 4772 |
| 868513545 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 83 | ... | <NA> | 42 | 207 | 76 | 4 | 38 | 528 | 280 | 204 | 4773 |
| 869214215 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 84 | ... | <NA> | 36 | 412 | 122 | 4 | 32 | 659 | 254 | 132 | 4774 |
| 870481685 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 85 | ... | <NA> | 40 | 318 | 128 | 3 | 37 | 537 | 285 | 157 | 4775 |
| 871728970 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 88 | ... | <NA> | 37 | 399 | 137 | 6 | 31 | 595 | 242 | 105 | 4776 |
| 872609185 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 89 | ... | <NA> | 40 | 407 | 133 | 2 | 38 | 589 | 277 | 144 | 4777 |
| 873449838 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 90 | ... | <NA> | 36 | 410 | 125 | 2 | 34 | 582 | 263 | 138 | 4778 |
| 874333572 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 91 | ... | <NA> | 37 | 361 | 101 | 4 | 33 | 620 | 249 | 148 | 4779 |
| 875020175 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 92 | ... | <NA> | 42 | 308 | 139 | 5 | 37 | 501 | 303 | 164 | 4780 |
| 876448528 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 96 | ... | <NA> | 38 | 285 | 106 | 7 | 31 | 559 | 258 | 152 | 4781 |
| 877855879 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 98 | ... | <NA> | 30 | 582 | 116 | 0 | 30 | 905 | 211 | 95 | 4782 |
| 878615867 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 99 | ... | <NA> | 31 | 533 | 104 | 2 | 29 | 864 | 221 | 117 | 4783 |
| 879854787 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 102 | ... | <NA> | 36 | 318 | 119 | 5 | 31 | 653 | 258 | 139 | 4784 |
| 880590869 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 103 | ... | <NA> | 34 | 515 | 148 | 7 | 27 | 651 | 251 | 103 | 4785 |
| 881210690 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 104 | ... | <NA> | 36 | 418 | 135 | 6 | 30 | 621 | 253 | 118 | 4786 |
| 882291265 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 105 | ... | <NA> | 31 | 623 | 141 | 4 | 27 | 841 | 219 | 78 | 4787 |
| 882736270 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 106 | ... | <NA> | 34 | 370 | 137 | 4 | 30 | 634 | 245 | 108 | 4788 |
| 883857883 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 109 | ... | <NA> | 39 | 342 | 139 | 6 | 33 | 567 | 260 | 121 | 4789 |
| 884605735 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 110 | ... | <NA> | 31 | 793 | 156 | 6 | 25 | 922 | 228 | 72 | 4790 |
| 885404024 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 111 | ... | <NA> | 36 | 603 | 182 | 3 | 33 | 762 | 243 | 61 | 4791 |
| 886382175 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 112 | ... | <NA> | 39 | 519 | 149 | 4 | 35 | 685 | 272 | 123 | 4792 |
| 888218684 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 116 | ... | <NA> | 39 | 207 | 127 | 7 | 32 | 605 | 267 | 140 | 4793 |
| 890021090 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 118 | ... | <NA> | 32 | 380 | 136 | 7 | 25 | 603 | 225 | 89 | 4794 |
| 893608687 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 123 | ... | <NA> | 35 | 265 | 96 | 4 | 31 | 872 | 249 | 153 | 4795 |
| 894495169 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 124 | ... | <NA> | 48 | 10 | 23 | 0 | 48 | 456 | 333 | 310 | 4796 |
| 895278567 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 125 | ... | <NA> | 44 | 166 | 68 | 1 | 43 | 540 | 302 | 234 | 4797 |
| 895809864 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 126 | ... | <NA> | 40 | 464 | 137 | 1 | 39 | 752 | 270 | 133 | 4798 |
| 896882463 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 127 | ... | <NA> | 37 | 655 | 181 | 5 | 32 | 695 | 260 | 79 | 4799 |
| 898665096 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 130 | ... | <NA> | 43 | 447 | 175 | 3 | 40 | 529 | 299 | 124 | 4800 |
| 898938920 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 131 | ... | <NA> | 43 | 281 | 115 | 1 | 42 | 485 | 312 | 197 | 4801 |
| 900227468 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 134 | ... | <NA> | 50 | 0 | 0 | 0 | 50 | 404 | 354 | 354 | 4802 |
| 901316636 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 137 | ... | <NA> | 43 | 133 | 79 | 3 | 40 | 503 | 283 | 204 | 4803 |
| 902211897 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 138 | ... | <NA> | 36 | 148 | 88 | 3 | 33 | 621 | 251 | 163 | 4804 |
| 902995627 | 902884228 | [1018027647, 1018027650, 1018027654, 101802765... | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 139 | ... | 2 | 31 | 273 | 113 | 8 | 23 | 587 | 239 | 126 | -999 |
| 903977177 | 903813946 | [1018027647, 1018027650, 1018027654, 101802765... | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 140 | ... | 2 | 52 | 0 | 0 | 0 | 52 | 406 | 354 | 354 | -999 |
| 904922800 | 904771513 | [1018027647, 1018027650, 1018027654, 101802765... | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 141 | ... | 2 | 33 | 214 | 104 | 8 | 25 | 569 | 247 | 143 | -999 |
| 906658064 | 906521029 | [1018027647, 1018027650, 1018027654, 101802765... | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 144 | ... | 2 | 28 | 206 | 81 | 8 | 20 | 623 | 203 | 122 | -999 |
| 907327499 | 907177554 | [1018027647, 1018027654, 1018027657, 101802766... | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 145 | ... | 2 | 52 | 0 | 0 | 0 | 52 | 405 | 353 | 353 | -999 |
| 908122381 | 907991198 | [1018027650, 1018027654, 1018027657, 1018027663] | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 146 | ... | 2 | 30 | 332 | 136 | 2 | 28 | 713 | 204 | 68 | -999 |
| 912640306 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 155 | ... | <NA> | 48 | 0 | 1 | 0 | 48 | 459 | 332 | 331 | 4805 |
47 rows × 34 columns
For this mouse, we can see that it progressed through a series of training stages starting on 3/15/2019 in behavior training boxes BEH.B-Box3 and BEH.B-Box1.
On 4/1/2019, it reached the TRAINING_5_images_A_handoff_ready, which meant that it was ready for transition to an imaging rig as soon as space became available.
On 4/4/2019, it was transitioned to ophys rig CAM2P.3, where it then underwent three days of habituation without imaging. This is evidenced by the fact that the session type for 4/4/2019, 4/5/2019, and 4/8/2019 was OPHYS_0_images_A_habituation and there was no associated ophys_session_id.
The first day of imaging for this mouse was on 4/9/2019, with session_type = OPHYS_1_images_A.
Note that this mouse has two OPHYS_5_images_B_passive sessions, the first taken in order (immediately after OPHYS_4_images_B), and second taken at the end of the sequence. The first OPHYS_5_images_B_passive does not have an ophys_session_id associated with it. This is likely due to that first session failing to meet quality control standards and being excluded from the dataset. The second OPHYS_5_images_B_passive was likely a retake, taken after the first was identified as having been failed.
In general, ophys behavior sessions that do not have associated ophys_session_ids are sessions for which the ophys data has been removed do to failure to meet quality control standards.
Aggregate data across all behavior sessions for this mouse#
Here we will iterate over all sessions for this mouse, build a behavior_session_dict which will have one behavior session object for every session that this mouse performed, with the key being the behavior_session_id
Note that this could take many minutes to complete. For each session in our new table, this_mouse_table, we are loading the behavior session NWB file from AWS, opening it as a BehaviorSession object using the AllenSDK, and adding that object as an entry to the dictionary.
If your cache directory is set to a location on your local computer, the NWB file for each session will be downloaded and added to the cache. This can take some time. If you were to re-run this cell a second time, it would access your cached NWB files instead of downloading them from AWS, allowing it to run substantially faster.
It is important to note that we will only be loading the behavior data here, even for sessions that had corresponding 2-photon imaging data. The get_behavior_ophys_experiment method would be used to get behavior and ophys data for ophys sessions. See the OphysSessionData notebook for details.
When the below cell completes, all behavior sessions for this mouse will be held in memory in the behavior_session_dict dictionary.
behavior_session_ids = this_mouse_table.index.values
behavior_session_dict = {}
for behavior_session_id in behavior_session_ids:
behavior_session_dict[behavior_session_id] = cache.get_behavior_session(behavior_session_id)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.6.0-alpha because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'hdmf-common' version 1.5.1 because version 1.8.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'core' version 2.5.0 because version 2.7.0 is already loaded.
return func(args[0], **pargs)
/opt/envs/allensdk/lib/python3.10/site-packages/hdmf/utils.py:668: UserWarning: Ignoring cached namespace 'hdmf-experimental' version 0.2.0 because version 0.5.0 is already loaded.
return func(args[0], **pargs)
Examine attributes of one BehaviorSession#
Below we will give a brief overview of what data is available for each BehaviorSession
Let’s look at some of the attributes of the last “handoff ready session”
We can filter the full table to get the last TRAINING_5_images_A_handoff_ready session. This would have been the last training session before the animal was subsequently handed off to the ophys team, after which all sessions were performed on a two-photon microscope.
Each session_type is distinguished by what stimulus was shown and what stage of training or imaging the mouse was in.
To learn more about the details of each session_type, see the VISUAL BEHAVIOR TASK OVERVIEW.
# get the last "TRAINING_5_" session_type
behavior_session_id = this_mouse_table.query('session_type == "TRAINING_5_images_A_handoff_ready"').index[-1]
# note that the following is functionally equivalent if you find the syntax easier to read:
# behavior_session_id = this_mouse_table[this_mouse_table['session_type'] == "TRAINING_5_images_A_handoff_ready"].index[-1]
# get the BehaviorSession object for this behavior session from the dictionary we created earlier
behavior_session = behavior_session_dict[behavior_session_id]
# list all attributes of the BehaviorSession object
behavior_session.list_data_attributes_and_methods()
['behavior_session_id',
'eye_tracking',
'eye_tracking_rig_geometry',
'get_performance_metrics',
'get_reward_rate',
'get_rolling_performance_df',
'licks',
'metadata',
'raw_running_speed',
'rewards',
'running_speed',
'stimulus_presentations',
'stimulus_templates',
'stimulus_timestamps',
'task_parameters',
'trials']
Note that any attribute can be followed by a ? in a Jupyter Notebook to see the docstring. For example, running the cell below will make a frame appear at the bottom of your browser with the docstring for the running_speed attribute.
behavior_session = behavior_session_dict[behavior_session_id]
behavior_session.running_speed?
Metadata#
The metadata attribute is a dictionary containing information about the BehaviorSession being examined, including information about the mouse, like the full_genotype and information about the session, such as the session_type
behavior_session.metadata
{'equipment_name': 'BEH.G-Box5',
'sex': 'F',
'age_in_days': 125,
'stimulus_frame_rate': 60.0,
'session_type': 'TRAINING_5_images_A_handoff_ready',
'date_of_acquisition': datetime.datetime(2019, 6, 26, 14, 2, 19, 825000, tzinfo=tzutc()),
'reporter_line': 'Ai93(TITL-GCaMP6f)',
'cre_line': 'Slc17a7-IRES2-Cre',
'behavior_session_uuid': UUID('50d5b9e3-44c4-414e-9b14-9205764f85ce'),
'driver_line': ['Camk2a-tTA', 'Slc17a7-IRES2-Cre'],
'mouse_id': '456915',
'project_code': 'VisualBehaviorMultiscope',
'full_genotype': 'Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-GCaMP6f)/wt',
'behavior_session_id': 895278567}
Task parameters#
The task_parameters attribute contains information about the structure of the behavior task for that specific session.
Here we can see that the stimulus_duration_sec is 0.25 seconds and the blank_duration_sec is 0.5 seconds. This determines the inter-stimulus interval.
behavior_session_dict[behavior_session_id].task_parameters
{'auto_reward_volume': 0.005,
'blank_duration_sec': [0.5, 0.5],
'image_set': 'images_A',
'n_stimulus_frames': 72126,
'omitted_flash_fraction': nan,
'response_window_sec': [0.15, 0.75],
'reward_volume': 0.007,
'session_type': 'TRAINING_5_images_A_handoff_ready',
'stimulus': 'images',
'stimulus_distribution': 'geometric',
'stimulus_duration_sec': 0.25,
'stimulus_name': 'Natural_Images_Lum_Matched_set_training_2017',
'task': 'change detection'}
Stimulus presentations#
The stimulus_presentations table contains one entry for each stimulus that was presented during the session, along with important metadata including stimulus start_time, image_name, and whether the stimulus was an image change (is_change = True) or an image omission (omitted = True).
behavior_session.stimulus_presentations.head(5)
| stimulus_block | stimulus_block_name | image_index | image_name | movie_frame_index | duration | start_time | end_time | start_frame | end_frame | is_change | is_image_novel | omitted | movie_repeat | flashes_since_change | trials_id | is_sham_change | active | stimulus_name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| stimulus_presentations_id | |||||||||||||||||||
| 0 | 0 | initial_gray_screen_5min | -99 | NaN | -99 | 299.979568 | 0.000000 | 299.979568 | 0 | 17985 | False | <NA> | <NA> | -99 | 0 | -99 | False | False | spontaneous |
| 1 | 1 | change_detection_behavior | 0 | im065 | -99 | 0.250188 | 299.979568 | 300.229756 | 17985 | 18000 | False | False | False | -99 | 1 | 0 | False | True | Natural_Images_Lum_Matched_set_training_2017 |
| 2 | 1 | change_detection_behavior | 0 | im065 | -99 | 0.250165 | 300.730091 | 300.980255 | 18030 | 18045 | False | False | False | -99 | 2 | 0 | False | True | Natural_Images_Lum_Matched_set_training_2017 |
| 3 | 1 | change_detection_behavior | 0 | im065 | -99 | 0.250165 | 301.480599 | 301.730764 | 18075 | 18090 | False | False | False | -99 | 3 | 1 | False | True | Natural_Images_Lum_Matched_set_training_2017 |
| 4 | 1 | change_detection_behavior | 0 | im065 | -99 | 0.250172 | 302.231110 | 302.481282 | 18120 | 18135 | False | False | False | -99 | 4 | 1 | False | True | Natural_Images_Lum_Matched_set_training_2017 |
Trials#
While the stimulus_presentations table has one entry for each individual stimulus that was presented, the trials table contains one entry for each behavioral trial, which consists of a series of stimulus presentations and is defined by the change_time.
On a given trial, a change_time is selected from a geometric distribution between 4 and 12 flashes after the time of the last change or the last lick.
On go trials, the image identity will change at the selected change_time. If the mouse licks within the response window (see response_window_sec entry of the `task_parameters attribute), that is considered a hit and a reward will be delivered. If the mouse fails to lick after the change, the trial is considered a miss.
On catch trials, a change_time is drawn, but the image identity does not change. If the mouse licks within the reward window, this is a false alarm and no reward is delivered. Correctly withholding a lick is called a correct reject.
This definition of a catch trial is a conservative one, and only considers the non-change stimulus presentations that are drawn from the same distribution as the change times. A less restrictive definition could consider every non-change stimulus presentation as a catch trial, and the false alarm rate can be computed this way as well.
If the mouse licks prior to the scheduled change_time, the trial is aborted and starts over again, using the same change_time for up to 5 trials in a row. This is to discourage mice from licking frequently, as they have to wait until the change time to get a reward.
# look at 5 random trials
behavior_session.trials.sample(5)
| start_time | stop_time | initial_image_name | change_image_name | is_change | change_time | go | catch | lick_times | response_time | ... | reward_time | reward_volume | hit | false_alarm | miss | correct_reject | aborted | auto_rewarded | change_frame | trial_length | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| trials_id | |||||||||||||||||||||
| 526 | 3835.435958 | 3843.474894 | im061 | im065 | True | 3839.209719 | True | False | [] | NaN | ... | NaN | 0.000 | False | False | True | False | False | False | 230191 | 8.038936 |
| 5 | 332.998169 | 335.916861 | im061 | im061 | False | NaN | False | False | [335.59997318452224] | NaN | ... | NaN | 0.000 | False | False | False | False | True | False | -99 | 2.918693 |
| 150 | 1258.675863 | 1269.733601 | im063 | im069 | True | 1265.468373 | True | False | [1266.1978047247976, 1266.3145172796212, 1266.... | 1266.197805 | ... | 1266.197805 | 0.007 | True | False | False | False | False | False | 75874 | 11.057737 |
| 362 | 2687.820583 | 2695.092299 | im066 | im062 | True | 2690.843820 | True | False | [] | NaN | ... | NaN | 0.000 | False | False | True | False | False | False | 161337 | 7.271716 |
| 318 | 2394.499748 | 2403.305874 | im066 | im065 | True | 2399.024039 | True | False | [2401.054307902232, 2401.3044888656586, 2401.7... | 2401.054308 | ... | NaN | 0.000 | False | False | True | False | False | False | 143840 | 8.806126 |
5 rows × 21 columns
We can examine one trial in some detail. Let’s randomly select a hit trial.
Some things to note:
The trial started at 831.2635398912244 seconds (
start_time) relative to the start of the session.The stimulus changed from ‘im063’ (
initial_image_name) to ‘im069’ (change_image_name) at t = 834.287206646593 seconds (change_time) relative to the start of the session.The animal’s first lick (
lick_times[0]) andresponse_timewas at t = 834.69975263 seconds relative to the start of the session.The
response_latency, which isresponse_time-change_time, was 0.41254598174464263 seconds.A reward (
reward_time) was delivered at 834.6997526283376 seconds relative to the start of the session. This was coincident with the first lick.
behavior_session.trials.query('hit').sample(random_state=0).to_dict('records')
[{'start_time': 1500.1606072620489,
'stop_time': 1507.465672607068,
'initial_image_name': 'im063',
'change_image_name': 'im065',
'is_change': True,
'change_time': 1503.2172430158384,
'go': True,
'catch': False,
'lick_times': array([1503.82983581, 1503.94659906, 1504.38021432, 1504.49697175,
1504.69711508, 1504.83052033, 1504.93062829, 1505.04734998,
1505.16410437, 1505.29752734, 1505.41427951, 1505.53103168,
1505.66445078, 1505.78121763, 1506.03137311, 1506.14812029,
1506.28154964, 1506.49838095, 1506.61511069, 1506.73186202]),
'response_time': 1503.8298358130269,
'response_latency': 0.6125927971884266,
'reward_time': 1503.8298358130269,
'reward_volume': 0.007,
'hit': True,
'false_alarm': False,
'miss': False,
'correct_reject': False,
'aborted': False,
'auto_rewarded': False,
'change_frame': 90129,
'trial_length': 7.305065345019102}]
The trials table includes the times of licks and rewards during each trial, but this information is also available in separate tables, described below.
Lick times#
The licks attribute is a dataframe with one entry for every detected lick onset time, assigned the time of the corresponding visual stimulus frame.
behavior_session.licks.sample(5)
| timestamps | frame | |
|---|---|---|
| 1654 | 2513.549276 | 150708 |
| 473 | 742.233067 | 44503 |
| 1557 | 2224.181202 | 133358 |
| 1741 | 2648.159639 | 158779 |
| 292 | 562.908287 | 33751 |
Rewards#
The rewards attribute is a dataframe containing one entry for every reward that was delivered, assigned the time of the corresponding visual stimulus frame. auto_rewarded is True if the reward was delivered without requiring a preceding lick. The first 5 change trials of each session are auto_rewarded = True.
behavior_session.rewards.sample(5)
| volume | timestamps | auto_rewarded | |
|---|---|---|---|
| 63 | 0.007 | 1780.672505 | False |
| 22 | 0.007 | 838.316614 | False |
| 72 | 0.007 | 2614.586260 | False |
| 6 | 0.007 | 391.622282 | False |
| 60 | 0.007 | 1676.883621 | False |
Running speed#
Mice are free to run on a circular disc during task performance. The running_speed table contains one entry for each read of the analog input line monitoring the encoder voltage, polled at ~60 Hz.
behavior_session.running_speed.head()
| timestamps | speed | |
|---|---|---|
| 0 | 0.000000 | 0.205226 |
| 1 | 0.016579 | 7.351836 |
| 2 | 0.033337 | 14.026109 |
| 3 | 0.049987 | 19.828231 |
| 4 | 0.068081 | 24.463707 |
Stimulus templates#
The stimulus_templates attribute is a dataframe containing one row per stimulus shown during the change detection task. The index is the image_name. The columns contain numpy arrays of the stimuli that were shown during the session.
The unwarped images represent the stimuli as they were seen by the mouse after correcting for distance from the mouse eye to the screen. The warped images show the exact image that was displayed on the stimulus monitor after spherical warping was applied to account for distance from the mouse’s eye.
behavior_session.stimulus_templates
| unwarped | warped | |
|---|---|---|
| image_name | ||
| im065 | [[nan, nan, nan, nan, nan, nan, nan, nan, nan,... | [[198, 197, 201, 207, 207, 202, 198, 201, 207,... |
| im077 | [[nan, nan, nan, nan, nan, nan, nan, nan, nan,... | [[129, 135, 140, 142, 140, 135, 129, 124, 117,... |
| im066 | [[nan, nan, nan, nan, nan, nan, nan, nan, nan,... | [[83, 85, 82, 78, 78, 81, 84, 84, 82, 82, 84, ... |
| im061 | [[nan, nan, nan, nan, nan, nan, nan, nan, nan,... | [[125, 126, 126, 126, 125, 125, 125, 125, 125,... |
| im063 | [[nan, nan, nan, nan, nan, nan, nan, nan, nan,... | [[105, 108, 107, 101, 97, 96, 98, 101, 103, 10... |
| im062 | [[nan, nan, nan, nan, nan, nan, nan, nan, nan,... | [[103, 103, 103, 105, 112, 125, 145, 172, 204,... |
| im085 | [[nan, nan, nan, nan, nan, nan, nan, nan, nan,... | [[54, 56, 50, 45, 45, 51, 56, 55, 51, 48, 49, ... |
| im069 | [[nan, nan, nan, nan, nan, nan, nan, nan, nan,... | [[251, 248, 255, 255, 255, 254, 246, 248, 255,... |
stimulus_templates = behavior_session.stimulus_templates.copy()
stimuli = stimulus_templates.index.values
plt.imshow(stimulus_templates.loc[stimuli[0]]['unwarped'], cmap='gray')
<matplotlib.image.AxesImage at 0x7f5c1c0b5bd0>
Stimulus timestamps#
Finally, a very important piece of information - the timestamps for each frame of visual stimulus display.
All behavioral measurements (running_speed, licks, & rewards) are made at the frequency of visual stimulus display (60Hz) and share frame times with the stimulus_presentations. You can use the frame index from any of the other behavior tables to determine the corresponding timestamp.
behavior_session.stimulus_timestamps
array([0.00000000e+00, 1.65791148e-02, 3.33371842e-02, ...,
4.50156656e+03, 4.50158323e+03, 4.50159990e+03])
Plot behavior data for a portion of one session#
Make some simple plotting functions to plot these datastreams
def plot_running(ax, behavior_session, initial_time, final_time):
'''
a simple function to plot running speed between two specified times on a specified axis
inputs:
ax: axis on which to plot
behavior_session: a behavior session object
initial_time: initial time to plot from
final_time: final time to plot to
'''
running_sample = behavior_session.running_speed.copy()
running_sample = running_sample[(running_sample.timestamps >= initial_time) &
(running_sample.timestamps <= final_time)]
ax.plot(running_sample['timestamps'],
running_sample['speed'])
def plot_licks(ax, behavior_session, initial_time, final_time):
'''
a simple function to plot licks as dots between two specified times on a specified axis
inputs:
ax: axis on which to plot
behavior_session: a behavior session object
initial_time: initial time to plot from
final_time: final time to plot to
'''
licking_sample = behavior_session.licks.copy()
licking_sample = licking_sample[(licking_sample.timestamps >= initial_time) &
(licking_sample.timestamps <= final_time)]
ax.plot(licking_sample['timestamps'], np.zeros_like(licking_sample['timestamps']),
marker = 'o', color = 'black', linestyle = 'none')
def plot_rewards(ax, behavior_session, initial_time, final_time):
'''
a simple function to plot rewards between two specified times as blue diamonds on a specified axis
inputs:
ax: axis on which to plot
behavior_session: a behavior session object
initial_time: initial time to plot from
final_time: final time to plot to
'''
rewards_sample = behavior_session.rewards.copy()
rewards_sample = rewards_sample[(rewards_sample.timestamps >= initial_time) &
(rewards_sample.timestamps <= final_time)]
ax.plot(rewards_sample['timestamps'], np.zeros_like(rewards_sample['timestamps']),
marker = 'd', color = 'blue', linestyle = 'none', markersize = 12, alpha = 0.5)
def plot_stimuli(ax, behavior_session, intial_time, final_time):
'''
a simple function to plot stimuli as colored vertical spans on a s
inputs:
ax: axis on which to plot
behavior_session: a behavior session object
intial_time: initial time to plot from
final_time: final time to plot to
'''
# get the stimulus presentations in the window we provided
stimulus_presentations_sample = behavior_session.stimulus_presentations.copy()
stimulus_presentations_sample = stimulus_presentations_sample[(stimulus_presentations_sample.end_time >= initial_time) &
(stimulus_presentations_sample.start_time <= final_time)]
# get a colormap for the different images
image_names = stimulus_presentations_sample[stimulus_presentations_sample.omitted==False].image_name.unique()
colors = sns.color_palette('hls', len(image_names))
# add white for omissions
image_names = np.hstack([image_names, 'omitted'])
colors = colors + [(1, 1, 1)]
# loop through images and plot them
for idx, stimulus in stimulus_presentations_sample.iterrows():
stim_color_ind = np.where(image_names==stimulus['image_name'])[0][0]
ax.axvspan(stimulus['start_time'], stimulus['end_time'], color=colors[stim_color_ind], alpha=0.25)
Select a time period during the session and generate the plot
initial_time = 775 # initial time for plot, in seconds
final_time = 800 # final time for plot, in seconds
plt.clf()
fig, ax = plt.subplots(figsize = (15,5))
plot_running(ax, behavior_session, initial_time, final_time)
plot_licks(ax, behavior_session, initial_time, final_time)
plot_rewards(ax, behavior_session, initial_time, final_time)
plot_stimuli(ax, behavior_session, initial_time, final_time)
ax.legend(['running speed', 'licks', 'rewards'])
ax.set_ylabel('running speed (cm/s)')
ax.set_xlabel('time in session (s)')
ax.set_xlim(initial_time, final_time)
ax.set_title('a short section of the session');
<Figure size 640x480 with 0 Axes>
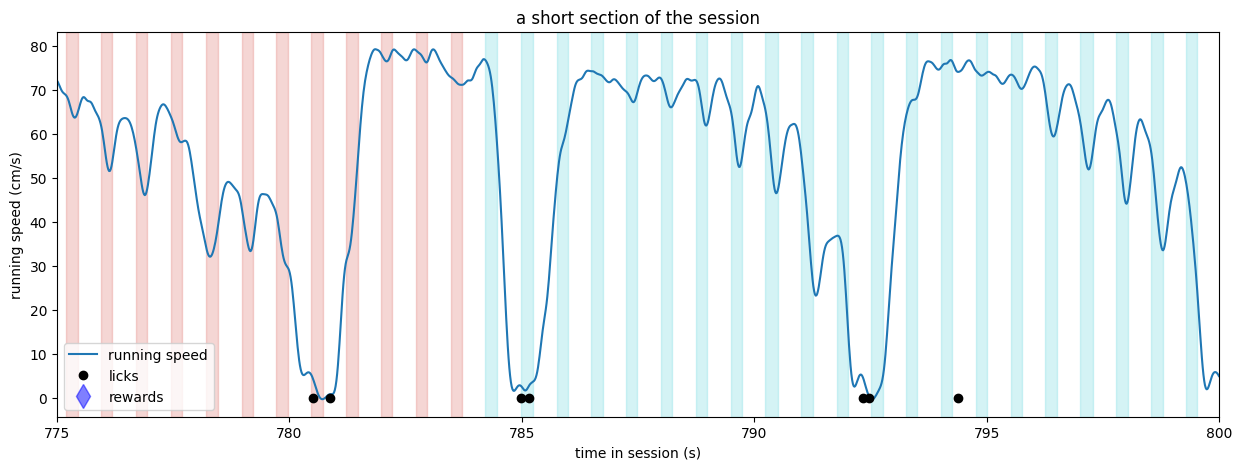
Above, we can see that stimuli were being delivered at a regular cadence (250 ms on, 500 ms off). There were changes to new stimuli at t = 778.6 and t = 793.7, as indicated by the change in the color of the bars. The mouse licked inside of the required response window following both stimulus changes and received a reward coincident with the first lick following the change. The subsequent licks are likely a result of the mouse consuming the water reward. There was also a brief bout of two licks, likely representing impulsivity, at t = 786.9.
Evaluate behavior performance across all sessions for this mouse#
One useful method of the BehaviorSession object is the get_performance_metrics method, which returns some summary metrics on the session.
# get behavior performance metrics for one session
behavior_session_dict[behavior_session_id].get_performance_metrics()
{'trial_count': 540,
'go_trial_count': 302,
'catch_trial_count': 44,
'hit_trial_count': 68,
'miss_trial_count': 234,
'false_alarm_trial_count': 1,
'correct_reject_trial_count': 43,
'auto_reward_count': 5,
'earned_reward_count': 68,
'total_reward_count': 73,
'total_reward_volume': 0.501,
'maximum_reward_rate': 4.553241861754873,
'engaged_trial_count': 212,
'mean_hit_rate': 0.2896157807630079,
'mean_hit_rate_uncorrected': 0.28757626472531317,
'mean_hit_rate_engaged': 0.4963845395179897,
'mean_false_alarm_rate': 0.08627629595837688,
'mean_false_alarm_rate_uncorrected': 0.027252588452010413,
'mean_false_alarm_rate_engaged': 0.1579499596448749,
'mean_dprime': 0.7145200008355794,
'mean_dprime_engaged': 1.0812555705834157,
'max_dprime': 1.5820865394018548,
'max_dprime_engaged': 1.5259011191946303}
You can also access performing metrics computed on a rolling basis across trials using the get_rolling_performance_df method
behavior_session.get_rolling_performance_df()
| reward_rate | hit_rate_raw | hit_rate | false_alarm_rate_raw | false_alarm_rate | rolling_dprime | |
|---|---|---|---|---|---|---|
| trials_id | ||||||
| 0 | NaN | NaN | NaN | NaN | NaN | NaN |
| 1 | NaN | NaN | NaN | NaN | NaN | NaN |
| 2 | NaN | NaN | NaN | NaN | NaN | NaN |
| 3 | NaN | NaN | NaN | NaN | NaN | NaN |
| 4 | NaN | NaN | NaN | NaN | NaN | NaN |
| ... | ... | ... | ... | ... | ... | ... |
| 535 | 0.0 | NaN | NaN | NaN | NaN | NaN |
| 536 | 0.0 | NaN | NaN | NaN | NaN | NaN |
| 537 | 0.0 | NaN | NaN | NaN | NaN | NaN |
| 538 | 0.0 | NaN | NaN | NaN | NaN | NaN |
| 539 | 0.0 | NaN | NaN | NaN | NaN | NaN |
540 rows × 6 columns
Aggregate performance metrics across sessions#
We can build out a new table that has all performance data for every session by iterating over the entries of our behavior_session_dict
This might take a minute or so. The AllenSDK will be extracting the performance data from the NWB file for every session individually.
# Let's use list comprehension to collect the metrics for each session
behavior_metrics_list = [behavior_session_dict[behavior_session_id].get_performance_metrics() for behavior_session_id in behavior_session_ids]
# Now turn it into a dataframe and set the index to be the behavior_session_id
behavior_performance_table = pd.DataFrame(behavior_metrics_list).set_index(behavior_session_ids)
behavior_performance_table.head()
| trial_count | go_trial_count | catch_trial_count | hit_trial_count | miss_trial_count | false_alarm_trial_count | correct_reject_trial_count | auto_reward_count | earned_reward_count | total_reward_count | ... | mean_hit_rate | mean_hit_rate_uncorrected | mean_hit_rate_engaged | mean_false_alarm_rate | mean_false_alarm_rate_uncorrected | mean_false_alarm_rate_engaged | mean_dprime | mean_dprime_engaged | max_dprime | max_dprime_engaged | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 862245493 | 118 | 0 | 0 | 0 | 0 | 0 | 0 | 118 | 0 | 118 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 863368489 | 1466 | 176 | 26 | 55 | 121 | 6 | 20 | 14 | 55 | 69 | ... | 0.459633 | 0.457263 | 0.818353 | 0.460983 | 0.522188 | 0.858631 | -0.104749 | -0.170745 | 0.562550 | 0.018189 |
| 863966974 | 1175 | 175 | 29 | 78 | 97 | 5 | 24 | 12 | 78 | 90 | ... | 0.597810 | 0.602154 | 0.803484 | 0.318760 | 0.327858 | 0.481352 | 0.844318 | 0.905205 | 1.260803 | 1.260803 |
| 864665204 | 786 | 246 | 39 | 86 | 160 | 7 | 32 | 16 | 86 | 102 | ... | 0.476640 | 0.479174 | 0.918062 | 0.219787 | 0.198738 | 0.440669 | 0.854237 | 1.678229 | 2.414661 | 2.414661 |
| 865131331 | 633 | 306 | 39 | 86 | 220 | 5 | 34 | 21 | 86 | 107 | ... | 0.401800 | 0.409642 | 0.955320 | 0.163136 | 0.110870 | 0.296478 | 0.770626 | 2.511776 | 3.393918 | 3.393918 |
5 rows × 23 columns
We can merge this table with the metadata table we built for this mouse because they have the same index (behavior_session_id)
this_mouse_table = this_mouse_table.merge(
behavior_performance_table,
left_index = True,
right_index = True)
this_mouse_table.head()
| ophys_session_id | ophys_container_id | mouse_id | indicator | full_genotype | driver_line | cre_line | reporter_line | sex | age_in_days | ... | mean_hit_rate | mean_hit_rate_uncorrected | mean_hit_rate_engaged | mean_false_alarm_rate | mean_false_alarm_rate_uncorrected | mean_false_alarm_rate_engaged | mean_dprime | mean_dprime_engaged | max_dprime | max_dprime_engaged | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| behavior_session_id | |||||||||||||||||||||
| 862245493 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 71 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 863368489 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 74 | ... | 0.459633 | 0.457263 | 0.818353 | 0.460983 | 0.522188 | 0.858631 | -0.104749 | -0.170745 | 0.562550 | 0.018189 |
| 863966974 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 75 | ... | 0.597810 | 0.602154 | 0.803484 | 0.318760 | 0.327858 | 0.481352 | 0.844318 | 0.905205 | 1.260803 | 1.260803 |
| 864665204 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 76 | ... | 0.476640 | 0.479174 | 0.918062 | 0.219787 | 0.198738 | 0.440669 | 0.854237 | 1.678229 | 2.414661 | 2.414661 |
| 865131331 | <NA> | NaN | 456915 | GCaMP6f | Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-G... | [Slc17a7-IRES2-Cre, Camk2a-tTA] | Slc17a7-IRES2-Cre | Ai93(TITL-GCaMP6f) | F | 77 | ... | 0.401800 | 0.409642 | 0.955320 | 0.163136 | 0.110870 | 0.296478 | 0.770626 | 2.511776 | 3.393918 | 3.393918 |
5 rows × 57 columns
Plot the max_dprime value for every session#
We can see that this particular mouse performed relatively consistently for every session as it progressed through training.
fig, ax = plt.subplots(figsize = (15,5))
ax.plot(np.arange(len(this_mouse_table)), this_mouse_table['max_dprime'], marker = 'o')
ax.set_xticks(range(len(this_mouse_table)))
ax.set_xticklabels(list(this_mouse_table['session_type'].values),rotation = 30, ha='right')
# make alternating black/gray vspans for visual clarity
colors = ['black', 'gray']
for ii in range(len(this_mouse_table)):
ax.axvspan(ii - 0.5, ii + 0.5, color = colors[ii%2], alpha=0.25)
ax.set_xlim(-0.5, len(this_mouse_table) - 0.5)
ax.set_ylabel('dprime')
ax.set_xlabel('session type')
ax.set_title("Max of rolling d' for every session for mouse {}".format(mouse_id))
fig.tight_layout()
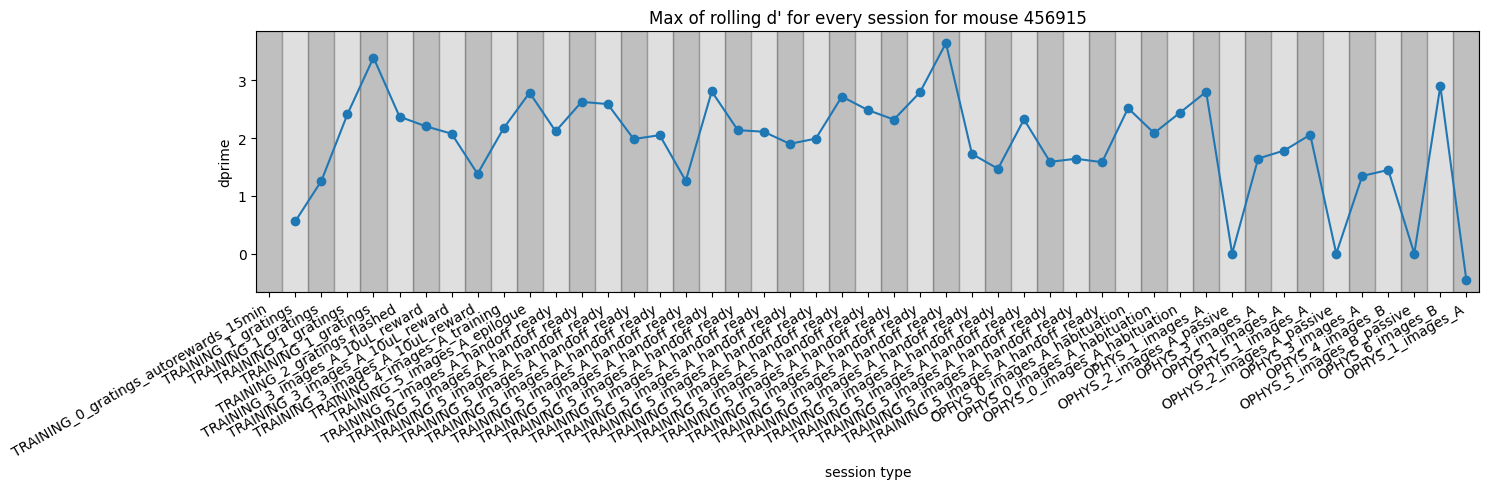
Note that the days with near zero dprime near the right side of the plot are all passive sessions where the lick spout was retracted and no rewards could be earned.
Exercises#
Can you color the points of the behavior performance metrics plot by some piece of metadata (one of the columns) in
this_mouse_table?How does the
max_dprimemetric compare with other metrics in therolling_performance_df?What does the performance look like for a different mouse?
How many sessions are there for each
session_type?Do all mice learn at a similar rate?
